Optimizing Algal Carbon Concentrating Mechanisms: Coordination of Biophysical and Biochemical Pathways for Enhanced Carbon Fixation
This article synthesizes current research on the coordination between biophysical and biochemical CO2 concentrating mechanisms (CCMs) in algae, a critical determinant of photosynthetic efficiency.
Optimizing Algal Carbon Concentrating Mechanisms: Coordination of Biophysical and Biochemical Pathways for Enhanced Carbon Fixation
Abstract
This article synthesizes current research on the coordination between biophysical and biochemical CO2 concentrating mechanisms (CCMs) in algae, a critical determinant of photosynthetic efficiency. Targeting researchers and biotechnology professionals, we explore the foundational principles of these mechanisms, their dynamic interplay under varying environmental conditions, and advanced methodologies for their study and manipulation. The review covers experimental approaches for assessing relative CCM contributions, discusses troubleshooting and optimization strategies to enhance carbon fixation, and validates findings through comparative analysis of model species and synthetic biology applications. By integrating foundational knowledge with cutting-edge methodological advances, this work aims to provide a comprehensive framework for leveraging algal CCMs to improve biofuel production, carbon sequestration technologies, and biomedical applications.
Fundamental Principles of Algal Carbon Concentrating Mechanisms: Structure, Function, and Environmental Regulation
Carbon Concentrating Mechanisms (CCMs) are vital adaptive strategies that enable photosynthetic organisms to overcome the limitations of Ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco), the key enzyme for carbon fixation. When environmental CO2 is limited, Rubisco's oxygenase activity competes with its carboxylase function, leading to photorespiration—a process that consumes energy and releases previously fixed carbon. CCMs actively increase the CO2 concentration around Rubisco's active site, thereby enhancing photosynthetic efficiency and reducing photorespiratory losses [1].
Two principal CCM types have evolved: biophysical CCMs rely on inorganic carbon transport and conversion through proteins and enzymes like carbonic anhydrases, while biochemical CCMs (such as C4 photosynthesis) utilize intermediate organic acids to shuttle carbon [1] [2]. In aquatic environments, where CO2 availability is particularly low, most microalgae and some macroalgae employ biophysical CCMs. However, research indicates that many species, including the green macroalga Ulva prolifera, operate both mechanisms in a complementary manner [2] [3] [4]. Understanding and optimizing the coordination between these systems represents a frontier in algal research with significant implications for biotechnology, climate change mitigation, and fundamental knowledge of global carbon cycling.
Core Concepts: Distinguishing the Two CCMs
What are the fundamental differences between biophysical and biochemical CCMs?
The table below summarizes the core distinctions between biophysical and biochemical carbon concentrating mechanisms.
Table 1: Fundamental Differences Between Biophysical and Biochemical CCMs
| Feature | Biophysical CCM | Biochemical CCM (C4-like) |
|---|---|---|
| Basic Principle | "Inorganic" mechanism; directly concentrates COâ‚‚ using transporters and compartmentalization [2]. | "Organic" mechanism; uses C4 acid intermediates to shuttle and release COâ‚‚ [2]. |
| Key Components | Carbonic anhydrases (CA), bicarbonate transporters (e.g., HLA3, LCIA), pyrenoid [5] [4]. | C4 enzymes: PEPC, PEPCK, PPDK [2] [4]. |
| Carbon Species Transported | Inorganic Carbon (COâ‚‚, HCO₃â») [2]. | Organic Carbon (C4 acids like malate, aspartate) [2]. |
| Energetic Cost | Consumes ATP for active transport of inorganic carbon [1]. | Consumes additional ATP/equivalent for the carboxylation-decarboxylation cycle. |
| Primary Function | Directly elevate COâ‚‚ at Rubisco site, suppressing photorespiration [1]. | Act as a biochemical pump to concentrate COâ‚‚ in specific cells or compartments. |
How do biophysical and biochemical CCMs coordinate in algae?
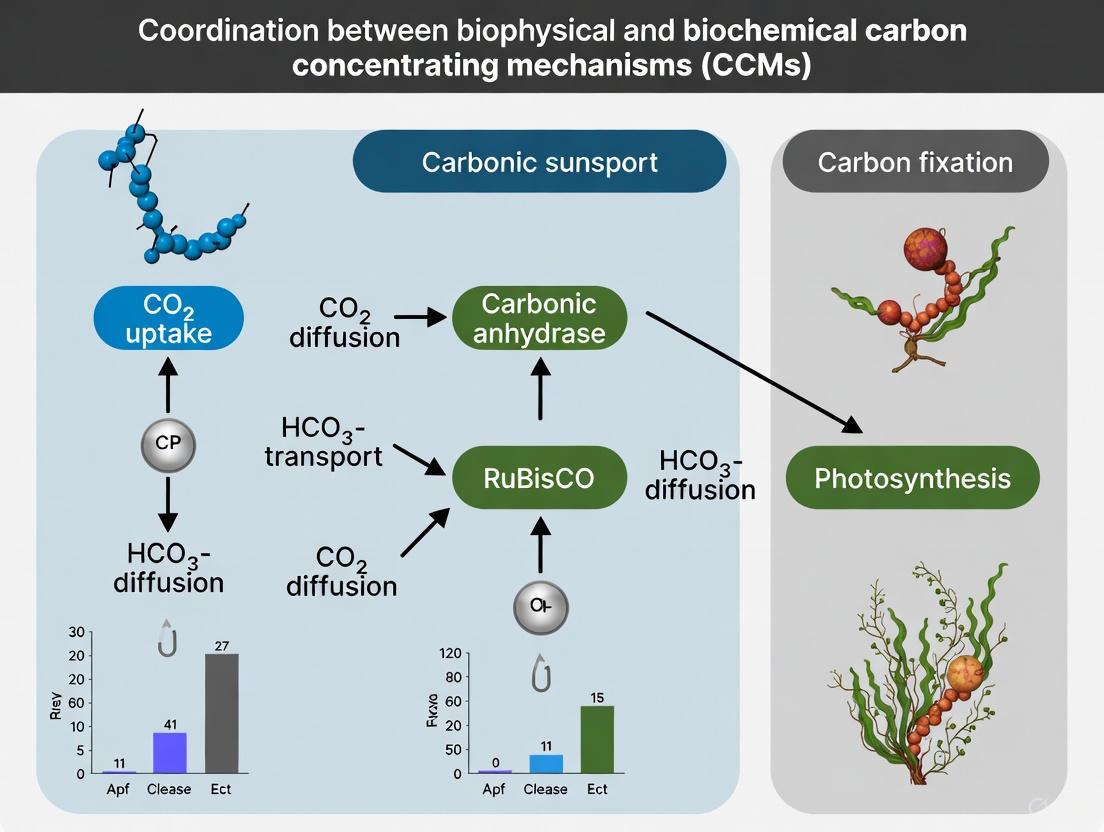
Research on the green macroalga Ulva prolifera reveals that its two CCMs do not operate in isolation but form a dynamic, complementary system. Inhibitor studies provide clear evidence of this coordination:
- When the biophysical CCM is inhibited by EZ (a carbonic anhydrase inhibitor), carbon fixation declines. Concurrently, cyclic electron flow around photosystem I increases, indicating the biochemical CCM becomes more active and can compensate for approximately 50% of the total carbon fixation [2] [6].
- Conversely, when the biochemical CCM is inhibited by MPA (a PEPCK inhibitor), the biophysical CCM is reinforced and can compensate for nearly 100% of the total carbon fixation [2] [6].
This plasticity allows U. prolifera to maintain high photosynthetic rates under fluctuating environmental conditions, contributing to its ability to form massive green tides [2]. The following diagram illustrates the coordinated workflow of these mechanisms and how researchers can probe them experimentally.
The Scientist's Toolkit: Essential Reagents and Methods
This section details key reagents, inhibitors, and model organisms used to dissect the functions of biophysical and biochemical CCMs.
Key Research Reagent Solutions
Table 2: Essential Reagents for CCM Research
| Reagent / Material | Function / Target | Experimental Application |
|---|---|---|
| Ethoxyzolamide (EZ) | Inhibits carbonic anhydrase (CA) activity [2]. | Suppresses the biophysical CCM. Used to assess its contribution to carbon fixation and to study compensatory mechanisms. |
| 3-Mercaptopicolinic Acid (MPA) | Inhibits phosphoenolpyruvate carboxykinase (PEPCK) [2]. | Suppresses the biochemical CCM. Used to quantify its role and the compensatory capacity of the biophysical CCM. |
| Acetazolamide (AZ) | Inhibits external, periplasmic carbonic anhydrase [2]. | Specifically targets the extracellular component of the biophysical CCM. |
| Chlamydomonas reinhardtii | Unicellular green alga with a well-characterized biophysical CCM and pyrenoid [5] [7] [8]. | A primary model organism for genetic studies, protein localization, and fundamental CCM research. |
| Ulva prolifera | Green macroalga known to operate both biophysical and biochemical CCMs [2] [3] [4]. | An ideal model for studying the coordination and environmental regulation of multiple CCMs. |
| Lucidone C | Lucidone C, CAS:102607-23-8, MF:C24H36O5, MW:404.5 g/mol | Chemical Reagent |
| Diminazene | Diminazene, CAS:536-71-0; 908-54-3, MF:C14H15N7, MW:281.32 g/mol | Chemical Reagent |
Quantitative Data from Key Experiments
The following table compiles critical quantitative findings from recent studies, providing a reference for expected experimental outcomes.
Table 3: Key Quantitative Findings in CCM Research
| Observation / Parameter | Quantitative Value | Context / Organism | Source |
|---|---|---|---|
| Biochemical CCM Compensation | ~50% of total carbon fixation | Contribution when biophysical CCM is inhibited in Ulva prolifera [2]. | |
| Biophysical CCM Compensation | ~100% of total carbon fixation | Capacity to compensate when biochemical CCM is inhibited in Ulva prolifera [2]. | |
| Km (CO₂) for U. prolifera | ~250 µM | Half-saturation constant for photosynthesis; much higher than ambient seawater CO₂ (5-25 µM) [4]. | |
| Critical PCOâ‚‚ for CCM Efficiency | As low as found in plants with biochemical CCMs (C4/CAM) | Level at which adding a biophysical CCM becomes energetically favorable in C3 plants [1]. |
Troubleshooting Common Experimental Challenges
Problem: Low carbon fixation efficiency in mutant algal strains.
- Question: How can I determine if the defect is in the CCM or another photosynthetic process?
- Solution:
- Measure photosynthetic Oâ‚‚ evolution or carbon fixation rates across a range of DIC concentrations to generate a P-C curve. An increased half-saturation constant (Kâ‚/â‚‚DIC) suggests impaired CCM activity [3] [4].
- Conduct experiments under photorespiratory (21% Oâ‚‚) and non-photorespiratory (2% Oâ‚‚) conditions. If the growth defect is rescued under low Oâ‚‚, the problem likely lies with the CCM's inability to suppress photorespiration effectively [7].
- Check for proper pyrenoid morphology and Rubisco localization. Mutants with disrupted pyrenoids (e.g., Chlamydomonas) show impaired fatty acid biosynthesis and carbon fixation, linking CCM function to other metabolic pathways [8].
Problem: Inconsistent results when using inhibitors like EZ and MPA.
- Question: Are my inhibitor concentrations and application methods correct?
- Solution:
- Optimal Concentrations: For Ulva prolifera, a final concentration of 50 µmol/L for EZ and 1.5 mmol/L for MPA is effective in culture experiments [2]. Always perform a gradient concentration detection for a new species or strain.
- Experimental Pre-treatment: Ensure algal samples are depleted of endogenous Ci sources before inhibitor application. This is typically done by transferring fragments to Ci-free, buffered artificial seawater for 30-60 minutes [2].
- Control Experiments: Use AZ (Acetazolamide) to distinguish between external (periplasmic) and total CA activity. AZ inhibits only external CA, while EZ penetrates cells and inhibits both external and internal CA [2].
Problem: Failed heterologous expression of algal CCM components in higher plants.
- Question: I have expressed a bicarbonate transporter in a C3 plant, but see no growth enhancement. What could be wrong?
- Solution:
- Check Subcellular Localization: Algal proteins may not automatically target to the correct compartment in plant cells. For example, the algal CAH6 and the putative Ci transporter LCI1 required fusion to an Arabidopsis chloroplast transit peptide for successful retargeting to the chloroplast [5] [9].
- Consider Component Stacking: Expression of individual Ci transporters (e.g., LCIA, HLA3) in Arabidopsis did not enhance growth. A fully functional CCM likely requires the synergistic expression of multiple components, including transporters, carbonic anhydrases, and a RuBisCO that can be packaged into a pyrenoid-like microcompartment [5] [9].
FAQs on CCM Function and Regulation
Q1: Why have biophysical CCMs not evolved more widely in land plants if they are so effective in algae?
- A1: Theoretical modeling suggests that the energy efficiency of a biophysical CCM in a typical C3 land plant leaf is highly dependent on factors like membrane permeability to CO₂. Under many conditions, the energetic cost of operating the CCM can be higher than the cost of photorespiration, making it unfavorable. The exception is in environments with very low CO₂ availability or low gas conductance, as seen in hornworts—the only land plants with a biophysical CCM [1].
Q2: Does the induction of a CCM completely suppress photorespiration in algae?
- A2: No, recent evidence challenges this long-held view. In Chlamydomonas, photorespiration remains active even when the CCM is operational under low COâ‚‚. Glycolate excretion and the down-regulation of glycolate dehydrogenase may serve as a safety valve to prevent the toxic accumulation of photorespiratory metabolites, indicating an integrated and dynamic relationship between CCM and photorespiration [7].
Q3: How do I know which CCM is dominant in the algal species I am studying?
- A3: The dominant mechanism can be environment-dependent. A combination of transcriptomics (looking for genes like HLA3, LCI1, PEPC, PEPCK), enzyme activity assays, and inhibitor studies (using EZ and MPA) is most effective. For example, in Ulva prolifera, the biophysical CCM is generally dominant, but the biochemical CCM can contribute significantly under specific stresses or when the biophysical CCM is impaired [2] [3].
Q4: Are there unexpected metabolic connections to the algal CCM?
- A4: Yes. Cutting-edge research shows that the pyrenoid, the core of the algal biophysical CCM, also plays a role in fatty acid biosynthesis. Under COâ‚‚ limitation, acetyl-CoA carboxylase (ACC) subunits localize to the pyrenoid periphery. Disrupting the pyrenoid impairs fatty acid and triacylglycerol biosynthesis, revealing a direct link between the CCM and carbon allocation to energy-rich storage compounds [8].
Frequently Asked Questions (FAQs)
Q1: My experiment shows negligible carbonic anhydrase (CA) activity in Chlamydomonas under low CO2, contrary to expectations. What could be wrong? The expression of carbonic anhydrases is highly specific. In Chlamydomonas, only a subset of CAs is strongly upregulated under low CO2. Transcriptomic studies reveal that among the twelve annotated CAs, primarily CAH1, CAH4, and CAH5 show significant induction (>2 fold-change) under low-CO2 conditions. Other isoforms may be downregulated or not induced. Confirm you are measuring the activity of the correct isoforms. Furthermore, note that a Cah1 mutant shows no apparent growth or photosynthetic phenotype, indicating it is not essential for the CCM, whereas a Cah3 mutant (thylakoid lumen-localized) is impaired in photosynthesis and exhibits a high-CO2-requiring phenotype [10].
Q2: I am observing inconsistent localization of introduced bicarbonate transporters in my transgenic plant model. How can I improve this? Retargeting algal components to appropriate organelles in higher plants can be challenging. A study expressing the algal plasma membrane transporter LCI1 in tobacco successfully redirected it to the chloroplast by fusing it to an Arabidopsis chloroplast transit peptide. Similarly, the chloroplastic carbonic anhydrase CAH6, which is naturally secreted in algae, was also retargeted to the chloroplast using the same strategy. Ensure your construct includes a validated transit peptide for your target organism and organelle [9].
Q3: How can I experimentally distinguish the activity of a C4-like pathway from the biophysical CCM in my algal cultures? The two mechanisms can be disentangled using enzyme-specific inhibitors and controlled conditions.
- Inhibitor Studies: Use the phosphoenolpyruvate carboxylase (PEPCase)-specific inhibitor 3,3-dichloro-2-dihydroxyphosphinoylmethyl-2-propenoate (DCDP). In the diatom Thalassiosira weissflogii, DCDP inhibition led to a >90% decrease in photosynthesis at low CO2, which was rescued by elevated CO2 or low O2, linking PEPCase activity to CO2 supply. This effect was minimal in the C3 green alga Chlamydomonas sp., where PEPCase serves a primarily anaplerotic role [11].
- Environmental Response: Monitor enzyme activity under varying light and CO2. Research on Ulva prolifera showed that key C3 enzyme (Rubisco) activity peaks in the morning, while C4 enzymes (PEPCase, PEPCKase) peak at noon under high irradiance, indicating a light-dependent role for the C4 pathway [12].
Q4: Why is my Chlamydomonas lci20 mutant showing a growth defect only during a sudden shift to very low CO2, but not when pre-acclimated? The LCI20 protein is a chloroplast envelope glutamate/malate transporter integral to photorespiration. During a sudden, severe CO2 limitation, the coordination between the rapidly induced CCM and photorespiratory metabolism becomes critical. LCI20 is proposed to supply amino groups for the mitochondrial conversion of glyoxylate to glycine. If this exchange is disrupted, photorespiratory metabolites can accumulate to toxic levels, impairing growth. In pre-acclimated cells, other compensatory mechanisms may be up-regulated to mitigate this defect [7].
Troubleshooting Guides
Diagnosing Issues with Bicarbonate Transport Assays
| Symptom | Potential Cause | Recommended Action |
|---|---|---|
| No HCO3- uptake detected in heterologous system (e.g., Xenopus oocytes). | Transporter not correctly localized to plasma membrane. | Confirm membrane localization with immunocytochemistry. For plant transporters, use an oocyte system validated for plant membrane proteins. |
| The expressed protein is a channel, not an active transporter. | Perform electrophysiology measurements to detect passive, channel-mediated flux. | |
| Inconsistent H14CO3- uptake rates in algal cultures. | Energy supply to the transporter is compromised. | Ensure cultures are well-lit; consider that cyclic electron flow (CEF) and mitochondrial respiration are key energy sources for transporters like HLA3 [13]. |
| The specific transporter is not induced. | Confirm the culture is properly acclimated to low CO2 conditions and check transcript levels of the target transporter (e.g., HLA3, LCIA). |
Resolving Problems in C4 Enzyme Activity Measurements
| Symptom | Potential Cause | Recommended Action |
|---|---|---|
| Low or no detectable PEPCase/PEPCKase activity in algal cell extracts. | Enzyme instability during extraction. | Include protease inhibitors and stabilizing substrates (e.g., PEP) in the extraction buffer. Perform extraction rapidly on ice. |
| Incorrect assumption of C4 pathway activity in the species. | Genomically verify the presence of key C4 enzymes (PEPCase, PEPCKase, PPDKase). Do not rely solely on PPDKase, as its role may be in photoprotection [12]. | |
| High background noise in radiometric assays. | Inefficient separation of metabolites. | Use validated separation methods like silicone oil centrifugation for short-term 14C uptake experiments [11]. |
Key Bicarbonate Transporters in Model Organisms
Table 1: Characteristics of selected bicarbonate transporters.
| Transporter | Organism | Gene | Function / Role | Key Characteristics / Localization |
|---|---|---|---|---|
| HLA3 | Chlamydomonas | Cre02.g097800 | Ci uptake into cytosol [9]. | ABC-type transporter, plasma membrane, induced under very low CO2 [9] [7]. |
| LCIA | Chlamydomonas | Cre06.g309000 | Ci transport from cytosol to stroma [9]. | Putative HCO3- channel, chloroplast envelope [9]. |
| BicA | Cyanobacteria | N/A | Na+-dependent HCO3- transporter [14]. | Low-affinity, high-flux, SulP family [14]. |
| SbtA | Cyanobacteria | N/A | Na+-dependent HCO3- transporter [14]. | High-affinity, low-flux [14]. |
| AE1 | Human | SLC4A1 | Cl-/HCO3- exchange [15]. | Acid loader; 911 amino acids, 13 transmembrane segments [15]. |
| NBCe1 | Human | SLC4A4 | Na+/HCO3- cotransport (electrogenic) [15]. | Acid extruder; 1035 amino acids [15]. |
Enzyme Activity Ranges in Algae
Table 2: Representative enzyme activities in algae under different conditions. NA: Data Not Available in the provided search results.
| Enzyme | Organism | Condition | Activity (nmol·minâ»Â¹Â·gFWâ»Â¹) | Notes |
|---|---|---|---|---|
| Rubisco (C3) | Ulva prolifera | Sunny day, 10:00 h | 274 | Activity drops significantly at noon [12]. |
| PEPCase (C4) | Ulva prolifera | Sunny day, noon | Peak Activity | Activity is high and correlates with irradiance [12]. |
| PEPCKase (C4) | Ulva prolifera | Sunny day | High | Significantly higher than on a cloudy day [12]. |
| PEPCase | T. weissflogii | Low CO2 (10 μM) | Functional | Inhibition causes >90% drop in photosynthesis [11]. |
Experimental Protocols
Protocol: Confirming HCO3- Transport Function inXenopusOocytes
Application: Functional characterization of putative bicarbonate transporters (e.g., LCIA, HLA3) [9]. Principle: The cRNA of the transporter is injected into oocytes. HCO3- uptake is quantified by measuring radioactivity (H14CO3-) in the oocytes after incubation.
Step-by-Step Methodology:
- Clone and Prepare cRNA: Clone the full-length coding sequence of the transporter gene into an oocyte expression vector (e.g., pOX). In vitro transcribe capped cRNA.
- Inject Oocytes: Defolliculate Stage V-VI Xenopus laevis oocytes. Inject 50 nL of cRNA (or water as a negative control) per oocyte. Incubate at 16°C in Barth's solution for 2-3 days to allow for protein expression.
- Uptake Assay: Wash oocytes in a CO2/HCO3--free buffer (e.g., ND96). Transfer groups of 10-15 oocytes to uptake solution (pH X.X) containing 1-2 µCi of H14CO3-.
- Terminate and Quantify: After a set time (e.g., 30-60 minutes), rapidly wash oocytes with ice-cold, non-radioactive uptake solution to stop the reaction. Lyse individual oocytes and quantify radioactivity via liquid scintillation counting.
- Data Analysis: Compare H14CO3- uptake in cRNA-injected oocytes to water-injected controls. Statistically significant increases confirm transport function.
Protocol: Assessing theIn VivoRole of C4 Metabolism using PEPCase Inhibition
Application: Determining the contribution of the C4 pathway to overall photosynthetic carbon fixation in diatoms and algae [11]. Principle: The specific PEPCase inhibitor DCDP is used to block the initial carboxylation step of the C4 pathway. The subsequent impact on photosynthesis is measured via O2 evolution.
Step-by-Step Methodology:
- Culture and Acclimate: Grow algal cells (e.g., Thalassiosira weissflogii) in air-equilibrated medium to acclimate them to low CO2 conditions.
- Inhibitor Incubation: Resuspend cell aliquots in fresh medium with or without the PEPCase inhibitor DCDP (e.g., 1 mM). Pre-incubate for a defined period (e.g., 30 minutes).
- Measure Photosynthesis: Transfer cell suspensions to an O2 electrode chamber. Illuminate at saturating light intensity and measure the rate of photosynthetic O2 evolution.
- Rescue Experiments: To confirm the effect is due to limited CO2 supply, repeat the inhibition assay under elevated CO2 (e.g., 150 µM) or low O2 (e.g., 80-180 µM) conditions. Recovery of photosynthesis under these conditions supports the role of PEPCase in a CCM.
- Interpretation: A strong suppression of O2 evolution by DCDP at low CO2 that is rescued by high CO2/low O2 indicates a major role for the C4 pathway in carbon accumulation.
Pathway and Workflow Visualizations
Carbon Concentration Mechanism Coordination
Diagram Title: Coordination of Biophysical and Biochemical CCM Components
Experimental Workflow for CCM Component Analysis
Diagram Title: Functional Analysis Workflow for CCM Components
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential reagents and their applications in algal CCM research.
| Reagent / Material | Function / Application | Specific Example / Note |
|---|---|---|
| DCDP (3,3-dichloro-2-dihydroxyphosphinoylmethyl-2-propenoate) | Specific inhibitor of phosphoenolpyruvate carboxylase (PEPCase). Used to dissect C4 metabolism contribution to photosynthesis [11]. | In T. weissflogii, 1 mM DCDP caused >90% decrease in O2 evolution at low CO2 [11]. |
| Mercaptopicolinic acid | Inhibitor of phosphoenolpyruvate carboxykinase (PEPCKase). Used to probe decarboxylation steps in C4 metabolism [11]. | Used in cell extraction solutions to inhibit PEPCKase during 14C uptake assays [11]. |
| H14CO3- | Radioactive tracer for measuring bicarbonate uptake and flux in various experimental systems (whole cells, oocytes) [9] [11]. | Provides direct measurement of transporter activity and carbon fixation pathways. |
| Chloroplast Transit Peptides | Protein sequences used to retarget algal proteins (e.g., transporters, CAs) to chloroplasts in plant transformation experiments [9]. | An Arabidopsis transit peptide successfully retargeted algal CAH6 and LCI1 to chloroplasts in tobacco [9]. |
| Antibodies (Specific) | Tools for detecting protein expression, localization (immunocytochemistry), and confirming knockout mutants. | An LCI20 antibody was used to confirm the absence of the protein in the lci20 Chlamydomonas mutant [7]. |
| Xenopus laevis Oocytes | Heterologous expression system for characterizing the function of ion transporters and channels [9]. | Validated for functional expression of algal Ci transporters like LCIA and HLA3 [9]. |
| Valtrate hydrine B4 | Valtrate hydrine B4, MF:C27H40O10, MW:524.6 g/mol | Chemical Reagent |
| Bph-742 | Bph-742, MF:C16H37O6P3, MW:418.38 g/mol | Chemical Reagent |
FAQs: Pyrenoid Function and Experimental Analysis
FAQ 1: What is the primary function of the pyrenoid? The pyrenoid is a phase-separated organelle found in the chloroplasts of most eukaryotic algae and hornworts. Its main function is to serve as the central hub for a biophysical CO2-concentrating mechanism (CCM) that enhances photosynthetic carbon assimilation. It concentrates CO2 around the enzyme Rubisco, thereby increasing its carboxylation rate and suppressing wasteful photorespiration [16] [17].
FAQ 2: My experiment shows low carbon fixation efficiency in a pyrenoid-deficient mutant. What is the underlying cause? This is an expected phenotype. The functional value of the pyrenoid matrix is to concentrate CO2 for Rubisco. Mutants with disrupted pyrenoids lack this concentrated CO2 supply, leading to increased Rubisco oxygenation and reduced carbon fixation efficiency. Research shows that preventing Rubisco condensation into a pyrenoid matrix carries a clear fitness cost [16].
FAQ 3: How can I experimentally distinguish the activity of a biophysical CCM from a biochemical CCM in my algal samples? You can use specific enzyme inhibitors in culture experiments to dissect the contributions of each mechanism.
- To inhibit the biophysical CCM: Use carbonic anhydrase (CA) inhibitors like Ethoxyzolamide (EZ) or Acetazolamide (AZ). These block the interconversion of HCO3- and CO2, which is crucial for biophysical carbon concentration [2].
- To inhibit the biochemical CCM: Use 3-mercaptopicolinic acid (MPA), an inhibitor of phosphoenolpyruvate carboxykinase (PEPCK), a key enzyme in the C4-like biochemical pathway [2]. The relative contribution of each CCM can be estimated by measuring changes in photosynthetic parameters (e.g., O2 evolution, carbon fixation rates) upon addition of these inhibitors [2].
FAQ 4: Why is a diffusion barrier considered critical for an efficient pyrenoid-based CCM? Computational models demonstrate that without a barrier to slow CO2 leakage, most CO2 generated within the pyrenoid escapes without being fixed by Rubisco. This creates a futile cycle where energy is wasted to concentrate CO2 that then diffuses away. The cell must expend additional energy to recapture this escaped CO2, making the CCM energetically inefficient. Models show that adding a diffusion barrier drastically reduces this leakage, enabling high Rubisco saturation (e.g., ~80%) at a lower energy cost (2-4 ATP per CO2 fixed) [18] [19].
FAQ 5: What are the known components of the pyrenoid in Chlamydomonas reinhardtii? The core molecular components in C. reinhardtii include:
- Rubisco: The main matrix component, assembled via interactions with EPYC1 [17].
- EPYC1 (Essential Pyrenoid Component 1): An intrinsically disordered linker protein essential for forming the phase-separated Rubisco matrix [17].
- SAGA1 and MITH1: Proteins necessary for forming the thylakoid membrane tubules that traverse the pyrenoid to deliver HCO3- [17].
- CAH3: A carbonic anhydrase located in the thylakoid lumen within the pyrenoid that converts HCO3- to CO2 [18].
- LCIB/LCIC: A protein complex forming a layer around the pyrenoid, hypothesized to recapture escaped CO2 [17].
- BST1-3: Putative HCO3- channels that allow HCO3- to move from the stroma into the thylakoid lumen [18].
Troubleshooting Guides
Problem: Inconsistent Pyrenoid Induction in Algal Cultures
Potential Causes and Solutions:
- Cause 1: Inadequate control of CO2 levels in the growth environment. The pyrenoid and CCM are highly inducible, primarily by low CO2 conditions [17].
- Solution: Pre-acclimate cultures to the target CO2 condition (e.g., high CO2 vs. air-level vs. very low CO2) for a sufficient period (≥24 hours). Use precise gas mixing systems or bubbled air with verified CO2 content for consistent results [20].
- Cause 2: Variations in light intensity during culture.
- Solution: Standardize light quantum levels and photoperiods across experiments, as light availability is a key regulator of photosynthesis and can influence CCM induction [2].
- Cause 3: Cell cycle effects. The pyrenoid partially dissolves and re-condenses during cell division [16] [17].
- Solution: Use synchronized cultures or sample cells at the same growth phase (e.g., mid-logarithmic phase) to minimize variability.
Problem: High CO2 Leakage in Pyrenoid Function Models
Potential Causes and Solutions:
- Cause: Lack of or defective diffusion barrier. Modeling indicates that a physical barrier is essential to reduce futile CO2 cycling [18] [19].
- Solution 1 (In vivo): In species like Chlamydomonas, ensure the integrity of the starch sheath, which can act as a diffusion barrier. Mutants with thinner or absent starch sheaths (e.g., sta2-1, sta11-1) show decreased CCM efficacy at low CO2 [19].
- Solution 2 (In silico): When constructing computational models, explicitly include a diffusion barrier with low permeability (on the order of 10â»â´ m/s) around the pyrenoid matrix to accurately simulate its function [19].
Problem: Difficulty in Distinguishing Between Multiple CCMs
Potential Causes and Solutions:
- Cause: Organisms can employ complementary CCMs. Some algae, like Ulva prolifera and Nannochloropsis oceanica, can operate both biophysical and biochemical (C4-like) CCMs, which may be coordinated or compensate for each other [2] [20].
- Solution: Implement a multi-omics approach. As demonstrated in N. oceanica, track transcriptomic, proteomic, and metabolomic profiles over time during a shift from high to low CO2. This can identify the coordinated upregulation of key genes and proteins specific to each CCM type, such as bicarbonate transporters/CAs for the biophysical CCM and PEPC/PEPCK for the biochemical CCM [20].
Data Presentation
Key Experimental Reagents for Pyrenoid and CCM Research
Table 1: Essential research reagents for studying pyrenoid-based carbon concentrating mechanisms.
| Reagent Name | Type | Primary Function in Experiments | Key Application/Note |
|---|---|---|---|
| Ethoxyzolamide (EZ) | Inhibitor | Inhibits carbonic anhydrase (CA) activity [2]. | Used to suppress the biophysical CCM; inhibits both extracellular and intracellular CA [2]. |
| Acetazolamide (AZ) | Inhibitor | Inhibits carbonic anhydrase (CA) activity [2]. | Used to suppress the biophysical CCM; primarily targets external periplasmic CA [2]. |
| 3-Mercaptopicolinic Acid (MPA) | Inhibitor | Inhibits phosphoenolpyruvate carboxykinase (PEPCK) [2]. | Used to suppress the biochemical C4-like CCM [2]. |
| EPYC1 Knockout Mutants | Genetic Tool | Lacks the linker protein essential for pyrenoid matrix formation in C. reinhardtii [17]. | Used to study pyrenoid assembly and the functional impact of a disrupted Rubisco matrix [17]. |
| Starch Sheath Mutants (e.g., sta2-1) | Genetic Tool | Possesses a thinner or absent pyrenoid starch sheath [19]. | Used to investigate the role of the starch sheath as a potential CO2 diffusion barrier [19]. |
Quantitative Metrics for PCCM Performance
Table 2: Key quantitative metrics used to evaluate the efficacy and efficiency of the pyrenoid-based CO2-concentrating mechanism (PCCM) from computational and experimental studies.
| Performance Metric | Description | Interpretation | Reported Values/Models |
|---|---|---|---|
| Rubisco Saturation | The extent to which Rubisco active sites are occupied by CO2, relative to the maximum possible rate [18]. | Directly measures PCCM efficacy. Higher saturation indicates a more effective CO2 concentration. | A model with an efficient PCCM can achieve ~80% saturation [19]. |
| ATP cost per COâ‚‚ fixed | The total number of ATP molecules consumed to fix one molecule of COâ‚‚ [18]. | Measures PCCM energetic efficiency. Lower cost is more efficient. | A passive-uptake PCCM can theoretically cost 2-3 ATP/COâ‚‚; an active mode costs 3-4 ATP/COâ‚‚ [19]. |
| Ci (Inorganic Carbon) Affinity | The ability of the cell to uptake Ci (COâ‚‚ + HCO₃â») from the external environment at low concentrations. | Indicates the activity of Ci uptake systems. Higher affinity is characteristic of an induced CCM. | Measured experimentally in mutants (e.g., starch sheath mutants show decreased affinity) and used to fit models [19]. |
Experimental Protocols
Protocol: Assessing CCM Contributions Using Specific Inhibitors
This protocol is adapted from studies on Ulva prolifera to distinguish the relative contributions of biophysical and biochemical CCMs to photosynthetic carbon fixation [2].
Workflow Overview:
Materials:
- Clark-type oxygen electrode system.
- Buffered artificial seawater (e.g., 20 mmol/L Hepes-NaOH, pH 8.0).
- Inhibitor stock solutions: 50 mmol/L Ethoxyzolamide (EZ) in DMSO, 1.5 mol/L 3-Mercaptopicolinic Acid (MPA) in DMSO.
- NaHCO₃ solution.
Step-by-Step Procedure:
- Culture Acclimation: Grow algal samples under the desired COâ‚‚ condition (e.g., high COâ‚‚ vs. air-level) for at least 24-48 hours to fully induce or repress the CCM.
- Ci Depletion: Cut algal samples into small fragments and transfer them to buffered artificial seawater that has been pre-aerated with Nâ‚‚ at low pH to remove dissolved COâ‚‚. Incubate for 30 minutes to deplete internal inorganic carbon pools.
- Inhibitor Application: Distribute the Ci-depleted samples into the reaction vessel of the O₂ electrode system. Add inhibitors to the respective treatment groups to achieve final concentrations of 50 μmol/L EZ (for biophysical CCM inhibition) or 1.5 mmol/L MPA (for biochemical CCM inhibition). Include a control group with an equivalent volume of solvent (DMSO).
- Measurement: Add NaHCO₃ to all samples to a final concentration of 2 mmol/L. Measure the steady-state rate of photosynthetic Oâ‚‚ evolution under saturating light (e.g., 200 μmol photons mâ»Â² sâ»Â¹).
- Data Analysis: Calculate the percentage inhibition for each CCM pathway using the formula:
% Inhibition = 100 x [1 - (Rate with inhibitor / Rate without inhibitor)]. The results indicate the relative contribution of each CCM to total carbon fixation under the tested conditions [2].
Protocol: Multi-Omics Analysis for CCM Identification
This protocol outlines a systems biology approach to identify components of multiple CCMs, as demonstrated in Nannochloropsis oceanica [20].
Workflow Overview:
Step-by-Step Procedure:
- Environmental Shift and Sampling: Grow a batch culture under high COâ‚‚ (e.g., 50,000 ppm). Harvest a sample for the "0-hour" time point. Then, rapidly transfer the culture to a very low COâ‚‚ (VLC) environment (e.g., 100 ppm). Collect subsequent samples at multiple time points (e.g., 3, 6, 12, and 24 hours post-shift) [20].
- Multi-Omics Profiling:
- Transcriptomics: Extract mRNA and perform RNA-Seq for all time points under both HC and VLC conditions. Identify differentially expressed genes (DEGs) in VLC compared to HC [20].
- Proteomics: Analyze protein extracts using mass spectrometry. Identify proteins that show significant changes in abundance in response to VLC [20].
- Metabolomics: Analyze metabolite pools to track changes in intermediates of the Calvin cycle, photorespiration, and potential C4 acids [20].
- Integrated Data Analysis:
- Triangulation: Cross-reference transcriptomic, proteomic, and metabolomic datasets to identify robust, coordinated responses to VLC.
- Functional Annotation: Map the induced genes/proteins to specific CCM types:
- Biophysical CCM: Look for significant upregulation of genes encoding bicarbonate transporters (BCTs) and various carbonic anhydrases (CAs) [20].
- Biochemical CCM: Look for induction of key C4-like pathway enzymes (e.g., PEPC, PEPCK, malate enzyme) at both the transcript and protein levels, alongside accumulation of C4 acids [20].
- Basal CCM: Investigate the upregulation of genes related to photorespiration and mitochondrial CO2 recycling systems [20].
The Scientist's Toolkit
Table 3: Key research reagent solutions for pyrenoid and CCM research.
| Research Reagent / Material | Function / Application |
|---|---|
| Chlamydomonas reinhardtii Mutant Library | A resource for identifying genes essential for pyrenoid function, CCM, and related processes via screening of targeted or random mutants [16]. |
| Fluorescently Tagged Protein Lines | Strains with fluorescently tagged pyrenoid components (e.g., EPYC1, Rubisco, LCIB) for visualizing protein localization and pyrenoid dynamics in vivo [16]. |
| Pyrenoid Proteome & Proxiome Datasets | Comprehensive lists of proteins localized to the pyrenoid and their interaction networks, providing a basis for hypothesizing protein functions [16]. |
| Computational (Reaction-Diffusion) Model of the PCCM | A quantitative framework to test hypotheses about PCCM operation, predict the impact of perturbations, and guide engineering strategies [18] [19]. |
| Fosfazinomycin B | Fosfazinomycin B, MF:C10H23N6O6P, MW:354.30 g/mol |
| DHA Ceramide | DHA Ceramide, MF:C40H67NO3, MW:610.0 g/mol |
Frequently Asked Questions (FAQs) and Troubleshooting
FAQ 1: My algal cultures are not achieving the expected biomass yield despite adequate CO2 supplementation. What are the key environmental factors I should optimize?
- Answer: Biomass yield is a function of several interdependent environmental factors. Beyond CO2, you must systematically optimize light, temperature, and nutrients. Recent research using machine learning models has identified that CO2 concentration and pH are often the most influential factors, followed by light colour (wavelength) and temperature [21]. Ensure your cultivation system allows for independent control and monitoring of these parameters. Use the table below (Summary of Optimal Environmental Conditions for Algal Biomass and Lipids) as a starting point for optimization.
FAQ 2: How can I non-destructively monitor the physiological status of my algal cultures in real-time during CCM experiments?
- Answer: Chlorophyll a fluorescence is a powerful, non-invasive technique to probe the efficiency of photosystem II (PSII), which is directly linked to the photosynthetic electron transport chain and affected by CCM activity [22] [23]. Parameters such as the maximum quantum yield of PSII (
Fv/Fm) provide a sensitive measure of photochemical efficiency and can indicate stress long before changes in biomass are detectable. Pulse-Amplitude-Modulation (PAM) fluorometers are specifically designed for these measurements, even in ambient light conditions [23].
FAQ 3: I observe a discrepancy between high electron transport rates (inferred from fluorescence) and low carbon fixation rates. What does this indicate?
- Answer: This disconnect often points to processes that consume reducing power without fixing carbon. A primary suspect is the induction of photorespiration [22] [23]. This can occur when the CCM is not fully active or is overwhelmed, leading to oxygenase activity by Rubisco. To confirm, simultaneously measure chlorophyll fluorescence and gas exchange (CO2 fixation). An increased electron requirement per molecule of CO2 fixed is a clear indicator of photorespiratory activity [23].
FAQ 4: Why does the chlorophyll content of my algal samples vary significantly under different nutrient regimes, and how does this affect my biomass estimates?
- Answer: Chlorophyll content per cell is a dynamic photoacclimation parameter. Under low light, cells increase chlorophyll to harvest more energy. Crucially, nutrient availability, especially nitrogen, strongly modulates this, as chlorophyll molecules are nitrogen-rich [24]. Under nitrogen limitation, cellular chlorophyll decreases, meaning that chlorophyll concentration can be a poor proxy for actual algal carbon biomass [24]. Always consider the Chlorophyll-to-Carbon (Chl:C) ratio for accurate biomass estimation under varying nutrient and light conditions.
FAQ 5: What is the most critical step when measuring the Fv/Fm parameter to assess the maximal PSII efficiency?
- Answer: Proper dark adaptation is essential. Samples must be dark-adapted for a sufficient period (typically 15-30 minutes) to ensure all PSII reaction centers are fully "open" and non-photochemical quenching is relaxed [22] [23]. Failure to do so will result in an underestimation of
Fv/Fmand an incorrect diagnosis of your culture's physiological state.
The following table consolidates optimal environmental conditions for maximizing algal biomass and lipid production, as identified in experimental studies.
Table 1: Summary of Optimal Environmental Conditions for Algal Biomass and Lipids
| Environmental Factor | Optimal Condition for Biomass/Lipids | Observed Effect / Quantitative Outcome | Key Genera Studied |
|---|---|---|---|
| COâ‚‚ Concentration | 9% | Significant positive correlation with lipid content (7.2–24.5%) and biomass (0.2–2.1 g Lâ»Â¹) [21]. | Chlorella, Botryococcus, Chlamydomonas, Tetraselmis, Closterium [21] |
| pH | 7 (Neutral) | Identified as a top-tier influential factor for growth and biochemical composition [21]. | Chlorella, Botryococcus, Chlamydomonas, Tetraselmis, Closterium [21] |
| Light Colour (Wavelength) | Red LED | Promoted the best growth rates in algal cultures [21] [25]. | Chlorella spp. and Chondracanthus acicularis (red seaweed) [21] [25] |
| Light Intensity | 3000 lux | Optimized biomass production under laboratory conditions [21]. | Chlorella, Botryococcus, Chlamydomonas, Tetraselmis, Closterium [21] |
| Temperature | 30°C | Supported optimal growth and biochemical productivity [21]. | Chlorella, Botryococcus, Chlamydomonas, Tetraselmis, Closterium [21] |
Essential Experimental Protocols
Protocol 1: ChlorophyllaFluorescence Measurement for PSII Health
This protocol is critical for non-invasively monitoring the photochemical performance of your algae, which is modulated by CCM activity [22] [23].
- Principle: The yield of chlorophyll fluorescence is inversely related to the photochemical efficiency of PSII. Measuring the fluorescence parameters provides insights into the energy partitioning within photosynthesis [23].
- Key Parameters:
- Fv/Fm: Maximum quantum yield of PSII. Values ~0.65-0.83 indicate healthy, unstressed phytoplankton. Lower values indicate photoinhibition or other stresses [23] [24].
- ΦPSII (Y(II)): Effective quantum yield of PSII under actinic (measuring) light. Estimates the rate of linear electron transport [23].
- Procedure:
- Dark Adaptation: Dark-adapt an algal sample for at least 15-20 minutes to ensure all reaction centers are open and relax non-photochemical quenching [22] [23].
- Measurement: Use a PAM fluorometer.
- Fâ‚€ Measurement: Apply a weak, modulated measuring beam to determine the minimum fluorescence yield.
- Fm Measurement: Apply a saturating pulse of light (e.g., 0.2-1s) to close all PSII reaction centers and measure the maximum fluorescence yield.
- Calculation: Calculate variable fluorescence
Fv = Fm - Fâ‚€and then the maximum quantum yieldFv/Fm[23]. - Light-Adapted Parameters: Under actinic light, you can further measure
Fm'(light-adapted maximum fluorescence) andFt(steady-state fluorescence) to calculateΦPSII = (Fm' - Ft) / Fm'[23].
Protocol 2: Machine Learning Workflow for Optimizing Environmental Factors
This modern approach efficiently navigates the complex, multi-factorial space of environmental modulators to optimize CCM performance and biomass yield [21].
- Principle: Machine learning (ML) models can analyze complex datasets from controlled experiments to predict the optimal combination of environmental factors for a desired output (e.g., high lipid content).
- Procedure:
- Factorial Experiment: Systematically culture algae across a matrix of different conditions (e.g., pH, temperature, COâ‚‚, light colour/intensity) [21].
- Data Collection: For each condition, quantify response variables such as biomass dry weight, lipid content (e.g., by gravimetric analysis after extraction), and protein content [21].
- Model Training: Use the experimental data to train ML models, such as Random Forest (RF), which has shown strong performance in this domain [21].
- Feature Importance Analysis: Use the trained model to identify which environmental factors (e.g., COâ‚‚, pH) are the most influential predictors of your target output [21].
- Validation: Validate the model's predictions with a new set of experiments conducted at the predicted optimal conditions.
Signaling and Regulatory Pathways
The following diagram summarizes the regulatory network through which key environmental signals modulate the Carbon Concentrating Mechanism (CCM) in algae, integrating both biophysical and biochemical components.
Diagram: Environmental Regulation of Algal CCM. Key environmental signals are transduced into coordinated gene expression and physiological changes, optimizing the coordination between biophysical and biochemical CCM components for enhanced carbon fixation and biomass yield.
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents and Materials for Algal CCM and Growth Studies
| Item | Function / Application in CCM Research | Example / Note |
|---|---|---|
| Bold's Basal Medium (BBM) | A standard synthetic nutrient medium for the axenic cultivation of a wide variety of freshwater algae [21]. | Provides essential macronutrients (N, P, S, Ca, Mg) and trace metals (Fe, Mn, Cu) necessary for growth [21]. |
| Provasoli's Enriched Seawater (PES) Medium | A common nutrient medium used for the cultivation of marine macroalgae and microalgae [25]. | Used in studies on red seaweeds like Chondracanthus acicularis; supports growth with a balanced nutrient profile [25]. |
| PAM Fluorometer | Measures chlorophyll a fluorescence parameters (e.g., Fv/Fm, ΦPSII, NPQ) to assess PSII photochemistry in vivo and non-destructively [22] [23]. |
Instruments like the IMAGING-PAM M-Series (Walz) allow for spatial resolution of photosynthetic performance [26]. Critical for monitoring culture health and CCM efficiency. |
| Pulse-Amplitude-Modulation (PAM) Technique | The underlying methodology that allows fluorescence measurement in ambient light by using a modulated measuring beam [23]. | This is the technological foundation that makes most modern fluorescence measurements possible outside of a dark lab. |
| COâ‚‚ Air Mixing System | To precisely control and maintain specific COâ‚‚ concentrations (e.g., 5%, 9%, 11%) in the culture aeration stream [21]. | Typically involves mass flow controllers and compressed air/COâ‚‚ gas tanks. Essential for studying CCM induction and its environmental modulation. |
| LED Light Panels (Multi-color) | To provide specific light wavelengths (e.g., red, blue, white) for studying the impact of light quality on photosynthesis and CCM activity [21] [25]. | Red light has been shown to promote optimal growth in several algal and seaweed species [21] [25]. |
| Nikkomycin Lx | Nikkomycin Lx, MF:C21H27N5O9, MW:493.5 g/mol | Chemical Reagent |
| Isariin D | Isariin D, MF:C26H45N5O7, MW:539.7 g/mol | Chemical Reagent |
In the field of photosynthetic research, Rubisco (Ribulose-1,5-bisphosphate carboxylase/oxygenase) presents a fundamental evolutionary puzzle. This key enzyme, responsible for carbon fixation during photosynthesis, is constrained by a well-documented inverse relationship between its specificity for COâ‚‚ versus Oâ‚‚ and its catalytic turnover rate. This trade-off directly impacts the efficiency of carbon fixation in photosynthetic organisms. For researchers engineering algal strains for enhanced carbon capture or biofuel production, understanding and navigating this trade-off is crucial for optimizing the coordination between biophysical and biochemical COâ‚‚ concentrating mechanisms (CCMs). This guide addresses the experimental challenges and solutions for investigating this critical relationship within the context of algal CCM optimization.
Key Concepts & Theoretical Framework
The Stability-Activity Trade-Off
Research on the adaptive evolution of Rubisco in C3 and C4 plants reveals that the enzyme's evolution has been constrained by stability-activity trade-offs [27]. The evolutionary shift from C3 to C4 photosynthesis involved a small number of mutations under positive selection that enhanced COâ‚‚ turnover rate at the cost of reduced COâ‚‚ specificity and structural stability [27].
- Molecular Mechanism: These adaptive mutations are often located near—but not directly in—the active site or at subunit interfaces, where they are inferred to enhance conformational flexibility required for the open-closed transition during catalysis [27].
- Evolutionary Preconditioning: The C3 to C4 transition was preceded by a period of increased Rubisco stability, creating the capacity to accept the necessary destabilizing mutations, followed by compensatory mutations that restored global stability [27].
Coordination of CCMs in Algal Systems
In aquatic environments where algae thrive, COâ‚‚ availability is limited, making CCMs essential for efficient photosynthesis. These mechanisms operate through complementary pathways:
- Biophysical CCMs: Rely on active transport of inorganic carbon (Ci) and the action of carbonic anhydrases (CAs) to concentrate COâ‚‚ near Rubisco [2].
- Biochemical CCMs: Similar to C4 photosynthesis in plants, they involve the fixation of Ci into C4 acid intermediates, which are later decarboxylated to release COâ‚‚ for Rubisco [2].
Recent studies demonstrate that these mechanisms operate jointly, with photorespiration remaining active even when CCMs are operational [7]. The relative contribution of each CCM type can shift based on environmental conditions, providing algae with metabolic plasticity [2].
Troubleshooting Common Experimental Challenges
FAQ: How can I determine the relative contributions of biophysical versus biochemical CCMs in my algal cultures?
- Challenge: Disentangling the simultaneous operation of multiple carbon fixation pathways.
- Solution: Use specific metabolic inhibitors in combination with measurements of photosynthetic output:
- Inhibit biophysical CCMs using ethoxyzolamide (EZ), a inhibitor of carbonic anhydrase activity [2].
- Inhibit biochemical CCMs using 3-mercaptopicolinic acid (MPA), a inhibitor of phosphoenolpyruvate carboxykinase (PEPCK) [2].
- Monitor changes in photosynthetic rate (e.g., via Oâ‚‚ evolution) and cyclic electron flow around photosystem I to quantify compensatory effects [2].
- Interpretation: In Ulva prolifera, when the biochemical CCM is inhibited, the biophysical CCM can compensate for almost 100% of carbon fixation, whereas when the biophysical CCM is inhibited, the biochemical CCM contributes approximately 50% [2]. This indicates dominance of the biophysical CCM with biochemical support.
FAQ: Why does my Rubisco engineering strategy lead to unstable enzymes despite improved kinetics?
- Challenge: Directly introducing mutations to enhance turnover rate often compromises structural stability.
- Solution: Implement a two-stage evolution strategy inspired by natural evolutionary paths:
- First, introduce stabilizing mutations to create a "robustness reservoir" [27].
- Then, introduce functionally necessary but destabilizing mutations that enhance conformational flexibility and turnover [27].
- Finally, select for compensatory secondary mutations that restore global stability without sacrificing the gained activity [27].
FAQ: How do I account for photorespiration when studying CCMs in algae?
- Challenge: The assumption that active CCMs completely suppress photorespiration is incorrect.
- Solution: Actively monitor photorespiratory metabolites and recognize that CCM and photorespiratory genes can be co-induced.
- Monitor glycolate excretion, as it indicates photorespiratory activity and helps prevent toxic metabolite accumulation [7].
- Use mutants defective in specific photorespiratory components (e.g., the LCI20 chloroplast envelope transporter) to dissect the relationship between CCM and photorespiration [7].
Essential Experimental Protocols
Protocol: Inhibitor-Based Dissection of CCM Contributions
This protocol determines the relative contributions of biophysical and biochemical CCMs by measuring photosynthetic Oâ‚‚ evolution under specific inhibition.
Workflow: Assessing CCM Contributions with Inhibitors
Materials & Reagents:
- Algal Culture (e.g., Ulva prolifera, Chlamydomonas reinhardtii)
- Clark-type Oâ‚‚ Electrode System (e.g., Hansatech) [2]
- Inhibitors:
- Buffered Artificial Seawater: 20 mmol/L Hepes-NaOH, pH 8.0 [2]
Procedure:
- Culture Preparation: Cut algal samples into uniform fragments (e.g., 1-cm for macroalgae) and culture in natural seawater for >2 hours prior to measurement [2].
- Ci Depletion: Transfer fragments to buffered artificial seawater without Ci for 30 minutes to deplete endogenous carbon sources. Use seawater previously aerated at low pH with pure Nâ‚‚ to remove COâ‚‚ [2].
- Inhibitor Application: Add inhibitors to buffered artificial seawater containing 2 mmol/L NaHCO₃ in four treatment conditions:
- Control (no inhibitors)
- EZ only (50 µM)
- MPA only (1.5 mM)
- EZ + MPA combination
- Oâ‚‚ Evolution Measurement: Measure photosynthetic Oâ‚‚ evolution rates at standardized conditions (e.g., 22°C, 200 μmol photons mâ»Â² sâ»Â¹) using an Oâ‚‚ electrode system [2].
- Data Analysis: Calculate percentage inhibition for each pathway:
- % Biophysical CCM contribution = [1 - (Rate with EZ)/(Control rate)] × 100
- % Biochemical CCM contribution = [1 - (Rate with MPA)/(Control rate)] × 100
Protocol: Measuring Photosynthetic Rates via Oâ‚‚ Production
This general protocol measures photosynthetic rate under different experimental conditions using respirometers.
Materials & Reagents:
- Respirometers with calibrated pipettes [28]
- Aquatic Plants (e.g., Elodea species) [28]
- Sodium Bicarbonate Solution (NaHCO₃) as carbon source [28]
- Light Source with adjustable intensity (portable lights) [28]
- Photometer for measuring light intensity (μmol mâ»Â² sâ»Â¹) [28]
Procedure:
- Plant Preparation: Obtain a 5-6 inch aquatic plant shoot, cut the basal end, and place it cut-end-up in a test tube [28].
- Setup: Fill the tube with sodium bicarbonate solution, cap with a rubber stopper holding a curved pipette, and ensure the pipette end is submerged. Seal with Parafilm [28].
- Control Preparation: Prepare a second tube without a plant as a thermobarometer to account for temperature and pressure changes [28].
- Equilibration: Suspend tubes in a water-filled beaker to buffer temperature changes. Wait ten minutes for equilibrium before starting measurements [28].
- Measurement: Turn light on and record fluid position in pipettes every two minutes for at least ten minutes. Reset using syringes if needed [28].
- Data Correction: Subtract thermobarometer readings from plant readings to control for atmospheric changes [28].
Research Reagent Solutions
Table 1: Essential Reagents for Investigating Rubisco Trade-offs and CCM Coordination
| Reagent/Equipment | Specific Function | Example Application | Considerations & References |
|---|---|---|---|
| Ethoxyzolamide (EZ) | Inhibits carbonic anhydrase (CA), blocking biophysical CCM | Quantifying biophysical CCM contribution by measuring O₂ evolution reduction after application | Use at 50 µM; inhibits both extracellular and intracellular CA [2] |
| 3-Mercaptopicolinic Acid (MPA) | Inhibits phosphoenolpyruvate carboxykinase (PEPCK), blocking biochemical CCM | Quantifying biochemical CCM contribution by measuring Oâ‚‚ evolution reduction after application | Use at 1.5 mM; specific to PEPCK-dependent C4 pathways [2] |
| Clark-type Oâ‚‚ Electrode | Measures photosynthetic Oâ‚‚ evolution rates | Core instrument for quantifying photosynthetic output under different inhibitor treatments or conditions | Standardize light (200 μmol photons mâ»Â² sâ»Â¹) and temperature (22°C) [2] |
| LCI20 Mutant Strains | Defective in chloroplast glutamate/malate transporter | Studying link between photorespiration, metabolite transport, and CCM operation under very low COâ‚‚ | Available from CLiP collection; shows impaired growth during sudden COâ‚‚ limitation [7] |
| Sodium Bicarbonate (NaHCO₃) | Provides dissolved inorganic carbon (DIC) for photosynthesis | Essential component in experimental media for maintaining carbon fixation in aquatic plants | Concentration should be optimized for specific organism; used in photosynthetic rate measurements [28] |
Data Presentation & Quantitative Analysis
Table 2: Quantitative Data on CCM Contributions from Inhibitor Studies in Ulva prolifera
| Experimental Condition | Photosynthetic Oâ‚‚ Evolution Rate | Inhibition Percentage | Compensatory Mechanism | Inferred Contribution to Carbon Fixation |
|---|---|---|---|---|
| Control (No inhibitor) | Baseline rate (100%) | 0% | N/A | Baseline carbon fixation |
| EZ (CA Inhibitor) | ~50% of baseline | ~50% | Increase in biochemical CCM activity | Biophysical CCM contributes ~50% |
| MPA (PEPCK Inhibitor) | ~50% of baseline | ~50% | Increase in biophysical CCM activity | Biochemical CCM contributes ~50% |
| EZ + MPA (Both inhibitors) | Strongly reduced | >90% | Limited compensation possible | Combined CCMs account for majority of fixation |
Key Findings: The data demonstrates the functional complementarity between CCM types. When one CCM is inhibited, the other can partially compensate, maintaining approximately 50% of photosynthetic activity [2]. This plasticity is crucial for algal survival in fluctuating environments.
Advanced Technical Considerations
Interorganellar Communication in CCM Function
Emerging research highlights the importance of organelle coordination in CCM operation:
The diagram illustrates how the LCI20 transporter, located in the chloroplast envelope, facilitates a malate/glutamate exchange that connects chloroplast metabolism with mitochondrial photorespiratory processes [7]. This exchange may supply amino groups for mitochondrial conversion of glyoxylate to glycine during photorespiration [7]. Simultaneously, mitochondria may supply ATP to power plasma membrane bicarbonate transporters like HLA3, creating an integrated energy network supporting CCM operation [7].
Pyrenoid as a Metabolic Hub
Recent findings in Chlamydomonas reinhardtii reveal that the pyrenoid serves beyond its traditional role in CCM:
- Fatty Acid Biosynthesis Center: Subunits of acetyl-CoA carboxylase (ACC) form phase-separated condensates at the pyrenoid periphery under COâ‚‚ limitation, linking carbon concentration to lipid metabolism [8].
- Metabolic Coordination: Pyrenoid-disrupted mutants show impaired fatty acid and triacylglycerol biosynthesis, supporting a model where CCMs supply carbon to FAS [8].
Advanced Methodologies for Investigating CCM Coordination: From Inhibitor Studies to Synthetic Biology
Carbon Concentrating Mechanisms (CCMs) are vital for efficient photosynthesis in algae, allowing them to overcome the slow diffusion of COâ‚‚ in water and the low affinity of Rubisco for COâ‚‚. Algae utilize two primary types of CCMs: biophysical CCMs, which actively transport and concentrate inorganic carbon (Ci) via carbonic anhydrases (CAs) and bicarbonate transporters, and biochemical CCMs (or C4-like pathways), which fix Ci into C4 organic acids before decarboxylation to supply COâ‚‚ to Rubisco [2]. Understanding the individual contribution of each mechanism is crucial for research on algal productivity, bloom dynamics, and bioengineering.
A powerful method to dissect these contributions involves the use of specific enzyme inhibitors. Ethoxyzolamide (EZ) inhibits carbonic anhydrase, thereby disrupting the biophysical CCM. Conversely, 3-Mercaptopicolinic acid (MPA) inhibits phosphoenolpyruvate carboxykinase (PEPCK), a key enzyme in the biochemical CCM [2]. By applying these inhibitors separately and in combination, researchers can quantify the relative roles and compensatory interactions between these two carbon fixation pathways.
Frequently Asked Questions (FAQs) & Troubleshooting
Q1: What are the specific functions of EZ and MPA in disrupting CCMs?
- EZ (Ethoxyzolamide): This cell-permeant inhibitor targets carbonic anhydrase (CA), both extracellular and intracellular [2]. CA catalyzes the interconversion between bicarbonate (HCO₃â») and COâ‚‚. By inhibiting CA, EZ disrupts the efficient conversion and transport of Ci forms, effectively impairing the biophysical CCM's ability to concentrate COâ‚‚ around Rubisco.
- MPA (3-Mercaptopicolinic Acid): This inhibitor specifically targets phosphoenolpyruvate carboxykinase (PEPCK) [2]. In the biochemical CCM of some algae, PEPCK is responsible for the decarboxylation of C4 acids (like oxaloacetate) to release COâ‚‚. Inhibiting PEPCK thus blocks the function of the C4-like biochemical pathway.
Q2: During inhibitor experiments, carbon fixation declines but is not completely eliminated. Why?
This is a common observation and is indicative of the robust, complementary coordination between biophysical and biochemical CCMs. Research on Ulva prolifera has shown that when the biophysical CCM is inhibited by EZ, the biochemical CCM can be reinforced and compensate for approximately 50% of the total carbon fixation [2]. Conversely, when the biochemical CCM is inhibited by MPA, the biophysical CCM can compensate for nearly 100% of carbon fixation [2]. This plasticity ensures that the alga maintains a baseline level of photosynthetic activity.
Q3: How do I determine the optimal concentration of EZ and MPA for my algal species?
The optimal concentration can vary based on the algal species and experimental conditions. It is critical to perform a dose-response curve. The following table summarizes concentrations used successfully in a study on Ulva prolifera [2]:
| Inhibitor | Target Enzyme | Mechanism Disrupted | Typical Working Concentration |
|---|---|---|---|
| EZ (Ethoxyzolamide) | Carbonic Anhydrase (CA) | Biophysical CCM | 50 µM [2] |
| MPA (3-Mercaptopicolinic Acid) | Phosphoenolpyruvate Carboxykinase (PEPCK) | Biochemical CCM | 1.5 mM [2] |
Troubleshooting Tip: If you observe no effect, verify the solubility and stability of your inhibitor stock solutions. If the inhibition effect is too severe, try a lower concentration and ensure you are measuring photosynthesis parameters (e.g., Oâ‚‚ evolution) within a linear range.
Q4: What are the expected changes in photosynthetic parameters when these inhibitors are applied?
The expected changes, based on the mechanism of action, are as follows:
| Parameter | EZ Application (Biophysical CCM inhibited) | MPA Application (Biochemical CCM inhibited) |
|---|---|---|
| Carbon Fixation Rate | Decreases [2] | Decreases [2] |
| Photosynthetic Oâ‚‚ Evolution | Decreases [2] | Decreases [2] |
| Compensatory CCM Activity | Biochemical CCM becomes more active [2] | Biophysical CCM becomes more active [2] |
| Cyclic Electron Flow (PSI) | Increases (supports reinforced biochemical CCM) [2] | May not show a significant change |
Q5: My results show unexpected activation of one CCM upon inhibition of the other. Is this normal?
Yes, this is a key finding and not an experimental error. The two CCMs are not independent but exist in a complementary coordination mechanism [2]. The inhibition of one pathway appears to trigger a compensatory upregulation of the other, a plasticity that allows the alga to maintain photosynthetic efficiency under fluctuating environmental conditions or chemical stress.
Diagram 1: Experimental workflow for assessing CCM contributions using inhibitors, showing the compensatory relationship between pathways.
Research Reagent Solutions
The following table lists key reagents essential for experiments designed to quantify CCM contributions using enzyme inhibition.
| Reagent | Function/Application in CCM Research |
|---|---|
| Ethoxyzolamide (EZ) | Cell-permeant inhibitor of carbonic anhydrase (CA); used to disrupt the biophysical CCM [2]. |
| 3-Mercaptopicolinic Acid (MPA) | Specific inhibitor of phosphoenolpyruvate carboxykinase (PEPCK); used to disrupt the biochemical (C4-like) CCM [2]. |
| Acetazolamide (AZ) | Specific inhibitor of external, periplasmic carbonic anhydrase; used to dissect internal vs. external CA activity [2]. |
| Buffered Artificial Seawater (e.g., with 20 mmol/L Hepes-NaOH, pH 8.0) | Provides a controlled ionic and pH environment for photosynthetic Oâ‚‚ evolution measurements after Ci depletion [2]. |
| Clark-type Oâ‚‚ Electrode System | Standard apparatus for measuring rates of photosynthetic oxygen evolution as a proxy for carbon fixation efficiency [2]. |
Experimental Protocols
Protocol: Assessing CCM Contributions via Inhibitors and Oâ‚‚ Evolution
This protocol is adapted from methods used in studies on Ulva prolifera [2].
Objective: To quantify the relative contributions of biophysical and biochemical CCMs to photosynthetic carbon fixation by applying specific enzyme inhibitors and measuring Oâ‚‚ evolution rates.
Materials:
- Healthy, axenic algal material.
- EZ inhibitor stock solution.
- MPA inhibitor stock solution.
- Buffered artificial seawater (e.g., 20 mM HEPES-NaOH, pH 8.0).
- Clark-type Oâ‚‚ electrode system.
- LED light source providing saturating quantum irradiance (e.g., 200 μmol photons mâ»Â² sâ»Â¹).
Procedure:
- Sample Preparation: Cut algal thalli into small, uniform fragments. Pre-culture them in natural seawater for several hours to ensure healthy metabolic activity.
- Ci Depletion: Transfer the fragments to Ci-free buffered artificial seawater. Bubble with pure Nâ‚‚ at low pH to remove dissolved COâ‚‚. Incubate for 30-60 minutes to deplete internal Ci pools.
- Inhibitor Incubation: Prepare experimental vials containing buffered artificial seawater with 2 mM NaHCO₃ as the Ci source.
- Control: No inhibitor added.
- +EZ: Add EZ to a final concentration of 50 µM.
- +MPA: Add MPA to a final concentration of 1.5 mM.
- (Optional) +EZ+MPA: Add both inhibitors to assess non-CCM background fixation. Equilibrate algal fragments in these inhibitor solutions for a predetermined time (e.g., 15-30 minutes) before measurement.
- Oâ‚‚ Evolution Measurement: Place the inhibitor-treated samples in the Oâ‚‚ electrode chamber. Measure the rate of photosynthetic Oâ‚‚ evolution under constant light and temperature.
- Data Analysis:
- Calculate the percentage inhibition for each treatment:
% Inhibition = 100 × [1 - (Rate with Inhibitor / Rate without Inhibitor)]. - The inhibition by EZ reflects the contribution of the biophysical CCM.
- The inhibition by MPA reflects the contribution of the biochemical CCM.
- The compensatory effect is evidenced by the remaining activity in each single-inhibitor treatment.
- Calculate the percentage inhibition for each treatment:
Diagram 2: EZ and MPA inhibition targets in biophysical and biochemical CCM pathways.
Data Presentation and Interpretation
The quantitative data from inhibition experiments can be clearly summarized in a table for easy comparison and interpretation.
Table: Example Quantitative Contributions of CCMs in Ulva prolifera under Inhibitor Treatment [2]
| Experimental Condition | Impact on Carbon Fixation | Inferred CCM Contribution | Key Compensatory Response |
|---|---|---|---|
| Control (No Inhibitor) | 100% baseline activity | Both CCMs operational | N/A |
| + 50 µM EZ (Biophysical CCM inhibited) | ~50% reduction | Biophysical CCM contributes ~50% | Biochemical CCM activity increases; Cyclic electron flow around PSI is enhanced [2]. |
| + 1.5 mM MPA (Biochemical CCM inhibited) | ~0% reduction (fully compensated) | Biochemical CCM contribution is compensated | Biophysical CCM is reinforced, compensating for nearly 100% of total carbon fixation [2]. |
| Theoretical: EZ + MPA | Severe reduction (>90%) | Confirms both pathways are major Ci acquisition routes | Little to no compensation possible. |
Interpretation Guide:
- Dominant Pathway: In Ulva prolifera, the biophysical CCM is the dominant pathway under standard conditions, as its inhibition leads to a direct and significant drop in carbon fixation.
- Supporting Role: The biochemical CCM plays a crucial supporting role, providing functional redundancy and resilience.
- Plasticity: The ability of one CCM to upregulate upon inhibition of the other highlights a sophisticated regulatory network for maintaining photosynthetic performance. This coordination is a key target for optimizing algal growth in both natural and biotechnological contexts.
Carbon Concentrating Mechanisms (CCMs) are essential biological systems that enhance photosynthetic efficiency by elevating the concentration of COâ‚‚ around the carbon-fixing enzyme RuBisCO. In algal systems, two primary CCM types exist: biophysical CCMs, which rely on inorganic carbon transport and conversion, and biochemical CCMs, which utilize organic carbon intermediates in C4-like pathways [2]. Understanding the coordination between these mechanisms requires metabolomic tracing, a powerful technique that uses stable isotope tracers to track carbon flux through metabolic pathways, providing dynamic insights into pathway activities [29] [30].
Frequently Asked Questions (FAQs)
Q1: What is the fundamental difference between metabolomics and metabolic tracing? Metabolomics provides a static snapshot of metabolite concentrations at a single point in time, showing what metabolites are present and their relative amounts. In contrast, metabolic tracing uses stable isotope labels to track how metabolites move through pathways over time, revealing the dynamic flow of carbon and the actual activity of metabolic pathways. While metabolomics might show that a metabolite pool has increased, only tracing can determine if this is due to increased production or decreased consumption [30].
Q2: Why is understanding both biophysical and biochemical CCMs important in algae research? Research on Ulva prolifera has demonstrated that these two CCM types function complementarily. When the biophysical CCM is inhibited, the biochemical CCM can compensate for approximately 50% of carbon fixation. Conversely, the biophysical CCM can compensate for nearly 100% of fixation when the biochemical CCM is inhibited. This plasticity allows algae to maintain efficient photosynthesis under varying environmental conditions [2].
Q3: What are the key advantages of stable isotopes over radioactive tracers? Stable isotopes (such as ¹³C, ¹âµN, ²H) are non-radioactive, making them safer to handle without special precautions. They allow parallel measurement of label incorporation into many downstream metabolites simultaneously using mass spectrometry or NMR. Modern instruments can detect these isotopes with high sensitivity, enabling comprehensive tracking through multiple pathways [29] [30].
Q4: What common issues affect labeling patterns in tracer experiments? Several factors can confound interpretation: insufficient tracer exposure time for slow-turnover pathways, failure to account for tracer dilution from endogenous sources, loss of labeled atoms as COâ‚‚ in decarboxylation reactions, and metabolic cross-talk between parallel pathways that can obscure the original tracer source [30].
Troubleshooting Guides
Poor Label Detection or Low Signal Intensity
| Problem | Potential Cause | Solution |
|---|---|---|
| Weak isotope signal | Tracer concentration too low | Optimize tracer dose through pilot experiments; ensure it doesn't disturb endogenous physiology [30] |
| Incomplete labeling | Incorrect exposure time for pathway kinetics | Extend labeling time for slower processes (e.g., protein synthesis vs. glycolytic lactate production) [30] |
| High variability between replicates | Inconsistent sample preparation or quenching | Standardize metabolite extraction protocols; use rapid quenching methods to halt metabolism instantly [29] |
| Carryover between samples | Contaminated LC-MS system | Include blank runs (mobile phase only) and solvent injections between samples to identify and reduce carryover [31] |
Data Quality and Interpretation Issues
| Problem | Potential Cause | Solution |
|---|---|---|
| Batch effects in large studies | Instrument drift across multiple batches | Use quality control (QC) samples from a pooled mixture of all samples; apply intra- and inter-batch normalization algorithms [31] |
| Misidentification of labeled peaks | Inaccurate isotopologue extraction | Use targeted extraction tools like MetTracer that generate theoretical m/z values for all possible isotopologues [32] |
| Inconsistent pathway interpretation | Natural isotope abundance not accounted for | Apply appropriate correction algorithms for natural abundance of ¹³C, ²H, ¹âµN, etc., before flux interpretation [29] |
| Unknown peaks in data | Limited metabolite database coverage | Use untargeted approaches with MS/MS spectral matching against public databases; report unknown compounds per Metabolomics Standards Initiative [33] |
Experimental Design Challenges
| Problem | Potential Cause | Solution |
|---|---|---|
| Unclear coordination between CCMs | Difficulty isolating mechanisms | Use specific inhibitors: EZ (ethoxyzolamide) for CA in biophysical CCMs; MPA (3-mercaptopicolinic acid) for PEPCK in biochemical CCMs [2] |
| Inability to distinguish nutrient sources | Single tracer limitations | Use multiple tracers simultaneously (e.g., [U-¹³C]-glucose, [U-¹³C]-glutamine, [U-¹³C]-acetate) with different labeling patterns [32] |
| Low coverage of labeled metabolites | Limited analytical scope | Implement global tracing technologies like MetTracer that combine untargeted metabolomics with targeted extraction for metabolome-wide coverage [32] |
Key Methodologies and Experimental Protocols
Protocol: Distinguishing Biophysical vs. Biochemical CCM Contributions
Purpose: Quantify the relative contributions of biophysical and biochemical CCMs to carbon fixation in algal systems [2].
Reagents and Materials:
- Ethoxyzolamide (EZ): Carbonic anhydrase inhibitor targeting biophysical CCM
- 3-mercaptopicolinic acid (MPA): PEPCK inhibitor targeting biochemical CCM
- Buffered artificial seawater (20 mmol/L Hepes-NaOH, pH 8.0)
- Clark-type Oâ‚‚ electrode system
- ¹³C-labeled bicarbonate or CO₂ source
Procedure:
- Prepare axenic cultures of algal species (e.g., Ulva prolifera).
- Divide cultures into four treatment groups: control, EZ-only (50 µM), MPA-only (1.5 mM), and EZ+MPA combination.
- Deplete endogenous carbon sources by incubating samples in Ci-free buffered seawater for 30 minutes.
- Add inhibitors to respective treatments 15 minutes before isotopic measurements.
- Introduce ¹³C-labeled carbon source (e.g., NaH¹³CO₃) simultaneously with photosynthetic O₂ evolution measurements.
- Terminate experiments at timed intervals (30, 60, 120 minutes) for metabolomic analysis.
- Quantify ¹³C incorporation into metabolites using LC-MS and calculate relative contributions of each CCM.
Protocol: Global Isotope Tracing with MetTracer
Purpose: Achieve comprehensive tracking of isotope labeling across the metabolome [32].
Workflow:
Critical Steps:
- Sample Preparation: Rapidly quench metabolism (liquid Nâ‚‚), extract metabolites with methanol:ethanol (1:1 v/v) containing internal standards.
- LC-MS Analysis: Use reverse-phase chromatography coupled to high-resolution mass spectrometer (Q-TOF or Orbitrap).
- Metabolite Annotation: Match MS2 spectra against standard libraries (e.g., HMDB, MassBank).
- Targeted Extraction: Generate theoretical m/z values for all possible isotopologues from annotated metabolites.
- Quantification: Extract and quantify isotopologue peaks, applying natural abundance correction.
- Pathway Mapping: Calculate labeling extents and patterns across metabolic pathways.
Research Reagent Solutions
Table: Essential Reagents for Metabolic Tracing in Algal CCM Research
| Reagent | Function | Application Example |
|---|---|---|
| [1,2-¹³C]glucose | Tracing glycolysis vs. PPP | Distinguishes oxidative PPP (M+1 lactate) from glycolysis (M+2 lactate) [29] |
| [U-¹³C]glutamine | Tracing TCA cycle & reductive carboxylation | Detects "backwards" TCA flux via M+5 citrate formation [29] |
| Ethoxyzolamide (EZ) | Inhibits carbonic anhydrase | Suppresses biophysical CCM by blocking HCO₃â»/COâ‚‚ interconversion [2] |
| 3-Mercaptopicolinic Acid (MPA) | Inhibits PEPCK | Suppresses biochemical CCM by blocking C4 acid decarboxylation [2] |
| Deuterated Internal Standards | Quality control & normalization | Monitors instrument performance; corrects technical variation [31] |
| ¹³C-bicarbonate (NaH¹³CO₃) | Direct carbon fixation tracing | Tracks inorganic carbon incorporation into metabolites [2] |
Data Analysis Pathway
Implementation Tools:
- XCMS, MZmine3: For peak detection and alignment [33]
- MetTracer, X13CMS: For isotopologue extraction and quantification [32]
- geoRge, mzMatch: For data normalization and batch effect correction [31] [33]
- PathVisio, MetaboAnalyst: For pathway mapping and visualization [33]
Advanced Applications: System-Wide Metabolic Coordination Analysis
The integration of global isotope tracing with computational modeling enables researchers to characterize system-wide metabolic homeostasis. Recent research in aging Drosophila demonstrates how this approach can reveal "a system-wide loss of metabolic coordinations" that impacts both intra- and inter-tissue metabolic homeostasis [32]. In algal studies, similar approaches can elucidate how environmental variations rewire carbon flux between biophysical and biochemical CCMs, informing strategies for optimizing biofuel production [2] [34].
For algal biofuel applications, Response Surface Methodology integrated with biokinetic modeling can optimize growth parameters including COâ‚‚ concentration (0.03-20%), light intensity (100-400 µE mâ»Â² sâ»Â¹), and nutrient ratios to maximize COâ‚‚ bio-fixation rates up to 1.2 g Lâ»Â¹ dâ»Â¹ and biomass productivity of 1.8 g Lâ»Â¹ [34].
Carbon Concentration Mechanisms (CCMs) are essential biological systems in algae that actively accumulate COâ‚‚ around the carbon-fixing enzyme Rubisco, thereby enhancing photosynthetic efficiency. Optimizing the coordination between biophysical CCMs (which use specialized protein pumps and compartments to concentrate COâ‚‚) and biochemical CCMs (which temporarily fix carbon into four-carbon compounds) represents a frontier in algal metabolic engineering [35]. Advanced genetic tools, particularly CRISPR-based systems, now enable precise modification of CCM components to boost carbon fixation, biomass productivity, and the yield of valuable biofuels and nutraceuticals [36].
This technical support center provides targeted troubleshooting and methodologies to help researchers overcome common challenges in CCM engineering projects.
The Scientist's Toolkit: Key Research Reagents
The table below details essential materials and their applications in algal genetic engineering experiments.
Table 1: Key Research Reagents for Algal Genetic Engineering
| Reagent / Tool Name | Primary Function | Example Application in CCM Research |
|---|---|---|
| CRISPR-Cas Systems [36] | Targeted gene knock-out, knock-in, and base editing | Disrupting genes for carbonic anhydrases to study their role in the biophysical CCM. |
| CRISPRa/i (dCas9) [36] | Precision gene activation or repression without DNA cleavage | Tunably overexpressing bicarbonate transporters or repressing photorespiration genes. |
| Base Editors (CBEs, ABEs) [36] | Single-nucleotide changes without double-strand breaks | Introducing point mutations in RuBisCO subunits to improve catalytic efficiency. |
| Antibiotic Resistance Markers [37] | Selection of successfully transformed algal cells | Using nourseothricin or paromomycin resistance to maintain engineered DNA constructs. |
| Ethoxyzolamide [35] | Inhibitor of biophysical CCM activity | Experimentally probing the contribution of biophysical CCMs to total carbon fixation. |
| 3-mercaptopicolinic acid [35] | Inhibitor of biochemical CCM activity | Selectively blocking biochemical CCM function to study compensatory mechanisms. |
| Anticancer agent 212 | Anticancer agent 212, MF:C19H12O3Te, MW:415.9 g/mol | Chemical Reagent |
| Phomarin | Phomarin, CAS:6866-87-1, MF:C15H10O4, MW:254.24 g/mol | Chemical Reagent |
Frequently Asked Questions (FAQs) & Troubleshooting
FAQ 1: Our CRISPR-Cas editing efficiency in a novel algal strain is very low. What are the primary factors to optimize?
Low editing efficiency is common in non-model algae. Focus on these key areas:
- Cas Protein and gRNA Expression: The codon usage of the Cas protein should be optimized for your specific algal host. Ensure robust expression by using strong, species-specific promoters (e.g., viral or endogenous constitutive promoters). For gRNA expression, empirical testing of RNA Pol III promoters (e.g., U6, tRNA) or ribozyme-flanked cassettes is often necessary [36].
- Delivery Method: The rigid cell wall of many algae is a major barrier.
- Electroporation is common but may require extensive optimization of voltage and pulse conditions.
- Biolistics (Particle Bombardment) is species-agnostic but can cause multi-copy integration and transgene silencing.
- Agrobacterium-mediated transformation or novel nanoparticle complexes (e.g., chitosan, polyethyleneimine) are emerging as promising alternatives for high-efficiency, low-copy delivery [36] [37].
- gRNA Design: Ensure your gRNA has high on-target activity and minimal predicted off-target effects. Use validated design tools and select target sites with high uniqueness in the genome [36].
FAQ 2: We have successfully engineered a CCM component, but the transformed algae show poor growth or no expected phenotypic improvement. How should we debug this?
This can result from metabolic burden or unintended system disruptions.
- Verify the Edit: Confirm the intended genetic modification using Sanger sequencing or next-generation sequencing. This rules out incomplete or off-target edits.
- Measure Physiological Impact: Quantify key photosynthetic parameters (e.g., Oâ‚‚ evolution, chlorophyll fluorescence) and carbon fixation rates directly. Inhibitors like ethoxyzolamide (biophysical CCM inhibitor) and 3-mercaptopicolinic acid (biochemical CCM inhibitor) can be used to dissect the specific contributions of each CCM sub-module and reveal compensatory mechanisms [35].
- Check for Metabolic Re-routing: Engineered algae may activate stress responses or re-route carbon flux. Use transcriptomics and metabolomics to profile global changes and identify unexpected bottlenecks or compensatory pathways [36].
FAQ 3: How can we safely cultivate algae with enhanced CCMs in outdoor settings, considering environmental release risks?
Implement robust biocontainment strategies as a mandatory safety layer.
- Synthetic Auxotrophy (Passive Strategy): Engineer the algae to be dependent on a nutrient not found in nature, such as phosphite, or an unnaturally high concentration of COâ‚‚. This can be achieved by knocking out native transporters for the standard nutrient (e.g., phosphate) while introducing a heterologous transporter for the novel nutrient [38].
- Active Kill-Switch Strategies: Design a genetic circuit where a lethal gene (e.g., a nuclease) is expressed in the absence of a synthetic signal molecule that is continuously supplied in the lab or bioreactor. If the algae escape, the signal is lost, triggering cell death [38].
Table 2: Troubleshooting Guide for Common Experimental Issues
| Problem | Potential Causes | Recommended Solutions |
|---|---|---|
| No transformants after selection. | Inefficient DNA delivery; ineffective antibiotic selection; toxic transgene. | Optimize delivery method parameters; validate antibiotic sensitivity; use inducible promoters for toxic genes [36] [37]. |
| High off-target mutation rate. | Low-fidelity Cas nuclease; gRNA with low specificity. | Switch to high-fidelity Cas variants (e.g., SpCas9-HF1); use computational tools to design more specific gRNAs; employ Cas12a for its reported lower off-target rates [36]. |
| Instability of the engineered trait. | Multi-copy integration causing silencing; loss of transgene over generations. | Use methods favoring single-copy integration (e.g., advanced Agrobacterium systems); include the transgene within the algal chromosome via homologous recombination where possible [37]. |
| Poor growth despite enhanced CCM. | Overloading of downstream metabolic pathways; metabolic burden. | Engineer the entire pathway coordinately (e.g., increase sink capacity for fixed carbon); use multiplexed CRISPR to edit multiple loci simultaneously [36]. |
Experimental Protocols for Key CCM Analyses
Protocol 1: Quantifying Relative Contributions of Biophysical and Biochemical CCMs
Objective: To determine the functional roles of biophysical and biochemical CCMs in a wild-type or genetically engineered algal strain.
Materials:
- Algal culture in mid-log growth phase.
- Carbon fixation assay system (e.g., IRGA - Infrared Gas Analyzer, or radioisotopic ¹â´C-bicarbonate).
- Specific inhibitors: Ethoxyzolamide (biophysical CCM inhibitor), 3-mercaptopicolinic acid (biochemical CCM inhibitor) [35].
Methodology:
- Divide the algal culture into four equal aliquots:
- Group A (Control): No inhibitor added.
- Group B (Biophysical inhibited): Treated with ethoxyzolamide.
- Group C (Biochemical inhibited): Treated with 3-mercaptopicolinic acid.
- Group D (Both inhibited): Treated with both inhibitors.
- Incubate the cultures with the inhibitors for a pre-optimized time (e.g., 30-60 minutes) under standard growth conditions.
- Measure the carbon fixation rate for each group using your chosen method (e.g., introducing ¹â´C-bicarbonate and quantifying incorporated radioactivity over a short time course).
- Data Analysis:
- The reduction in fixation in Group B represents the contribution of the biophysical CCM.
- The reduction in Group C represents the contribution of the biochemical CCM.
- The fixation rate in Group D indicates the baseline, CCM-independent fixation.
- Analyze coordination by observing if inhibition of one mechanism leads to a compensatory increase in the activity of the other [35].
Experimental Workflow for CCM Analysis
Protocol 2: Implementing a CRISPR-Based Base Edit in a CCM Gene
Objective: To introduce a specific point mutation in a gene of interest (e.g., a RuBisCO subunit) using a CRISPR-Cytosine Base Editor (CBE).
Materials:
- Plasmid DNA encoding a CBE (e.g., BE4max) and the specific gRNA.
- Delivery equipment (e.g., electroporator).
- Selective media (e.g., containing paromomycin).
- PCR and sequencing primers for genotyping [36].
Methodology:
- Design: Design the gRNA to bind ~15 nucleotides upstream of the target cytosine (within the editing window, typically positions 4-8).
- Construct Assembly: Clone the gRNA sequence into the CBE plasmid vector using standard molecular cloning techniques.
- Delivery: Introduce the plasmid into your algal strain via your optimized method (e.g., electroporation).
- Selection and Screening: Plate cells on selective media. After colonies appear, pick them and grow in 96-well deep plates for genotyping.
- Genotyping: Extract genomic DNA and perform PCR amplification of the target region. Send the PCR product for Sanger sequencing to confirm the intended C-to-T (or G-to-A on the opposite strand) base change [36].
Advanced Engineering: Pathways and Logic Gates
Engineering enhanced CCMs often requires coordinated expression of multiple genes. Advanced CRISPR tools like CRISPRa (activation) and CRISPRi (interference) allow for this multiplexed, tunable control. The diagram below conceptualizes a genetic circuit designed to dynamically optimize CCM function in response to internal metabolic cues.
Logic Gate for Dynamic CCM Optimization
Troubleshooting Guides & FAQs
Common Experimental Issues & Solutions
FAQ: My multi-omics data shows inconsistent results between genomic loci and protein expression. How should I proceed?
- Potential Cause: Post-transcriptional regulation or protein degradation mechanisms may be creating a disconnect between transcriptomic and proteomic data.
- Solution: Perform colocalization analysis, such as Mendelian Randomization, to establish whether a causal relationship exists between the genetic variant and the protein expression. Spatial proteomics can then validate whether the corresponding proteins are present and correctly localized [39] [40].
FAQ: I am observing high variability in carbon fixation efficiency in my algal cultures. What could be the reason?
- Potential Cause: Uncoordinated activity between biophysical and biochemical COâ‚‚ Concentration Mechanisms (CCMs). Environmental factors like light quality and COâ‚‚ concentration significantly influence which mechanism dominates [2] [41].
- Solution: Systematically inhibit specific CCM pathways using chemical inhibitors like EZ (for biophysical CCM) and MPA (for biochemical CCM) to quantify their individual contributions. Ensure culture conditions (COâ‚‚%, light, inoculation density) are strictly controlled and replicated [2].
FAQ: How can I distinguish between a biophysical CCM and a biochemical CCM in my algae strain?
- Solution: Use specific inhibitors in conjunction with measurements of photosynthetic Oâ‚‚ evolution or carbon fixation rates.
- Ethoxyzolamide (EZ): Inhibits carbonic anhydrase (CA), a key enzyme in the biophysical CCM.
- 3-Mercaptopicolinic Acid (MPA): Inhibits phosphoenolpyruvate carboxykinase (PEPCK), a key enzyme in the C4 biochemical CCM [2]. A decline in carbon fixation upon adding EZ indicates an active biophysical CCM, while a decline with MPA indicates an active biochemical CCM.
Quantitative Data on CCM Contributions
The table below summarizes experimental data on the relative contributions of CCMs in the green macroalga Ulva prolifera, as determined by inhibitor studies [2].
Table 1: Contribution of Biophysical and Biochemical CCMs to Carbon Fixation in Ulva prolifera
| Condition | Inhibitor Used | Target Pathway | Observed Effect on Carbon Fixation | Estimated Contribution |
|---|---|---|---|---|
| Inhibition of Biophysical CCM | Ethoxyzolamide (EZ) | Carbonic Anhydrase | Declined | Dominant (~100% compensation capacity) |
| Inhibition of Biochemical CCM | 3-mercaptopicolinic acid (MPA) | PEPCK | Declined | Supporting (~50% of total) |
Detailed Experimental Protocols
Protocol 1: Differentiating CCM Contributions using Inhibitors
This protocol is adapted from studies on Ulva prolifera to quantify the role of biophysical and biochemical CCMs [2].
- Sample Preparation: Take healthy algal thalli and cut them into uniform fragments (e.g., 1-cm length). Pre-culture them in buffered artificial seawater (e.g., 20 mmol/L Hepes-NaOH, pH 8.0) in the absence of inorganic carbon (Ci) for 30 minutes to deplete endogenous Ci sources.
- Inhibitor Preparation: Prepare stock solutions of the inhibitors.
- Ethoxyzolamide (EZ): A potent inhibitor of carbonic anhydrase (extracellular and intracellular).
- 3-mercaptopicolinic acid (MPA): An inhibitor of phosphoenolpyruvate carboxykinase (PEPCK).
- Experimental Setup: Set up the following treatments in buffered artificial seawater with 2 mmol/L NaHCO₃:
- Control: No inhibitors added.
- EZ Treatment: Add EZ to a final concentration of 50 µmol/L.
- MPA Treatment: Add MPA to a final concentration of 1.5 mmol/L.
- Measurement: Transfer the algal fragments to a Clark-type Oâ‚‚ electrode system. Measure the rate of photosynthetic Oâ‚‚ evolution at a controlled temperature (e.g., 22°C) and saturating quantum irradiance (e.g., 200 μmol photons mâ»Â² sâ»Â¹).
- Calculation: Calculate the percentage inhibition for each treatment using the formula:
Percentage Inhibition = 100 x [1 - (Rate with inhibitors / Rate without inhibitors)]
Protocol 2: An Integrated Multi-Omics Workflow for CCM Gene Validation
This protocol outlines a computational and experimental pipeline for identifying and validating candidate genes involved in CCM regulation, inspired by large-scale genetic studies [39].
Genomic Variant Identification:
- Perform a genome-wide association study (GWAS) meta-analysis to identify genetic variants (SNPs) associated with CCM-related traits.
- Annotate candidate genes for significant variants using both proximity (nearest gene) and functional prediction methods like Polygenic Priority Score (PoPS).
Transcriptomic & Proteomic Integration:
- Integrate the genomic loci with expression quantitative trait locus (eQTL) and protein quantitative trait locus (pQTL) data. This determines if the genetic variant is associated with changes in mRNA or protein levels of the candidate gene.
- Use Mendelian Randomization and colocalization analyses to assess potential causal relationships between the genetic variant, gene/protein expression, and the CCM trait.
Functional and Spatial Validation:
- Perform single-cell RNA sequencing to identify specific cell types (e.g., endothelial cells in vascular plants) that differentially express the candidate genes.
- Use spatial proteomics to confirm the subcellular localization and abundance of the proteins encoded by the candidate genes, bridging the gap between genetic association and functional protein activity [40].
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents for CCM and Multi-Omics Research
| Reagent / Material | Function / Application |
|---|---|
| Ethoxyzolamide (EZ) | A potent inhibitor of carbonic anhydrase (CA). Used to suppress the biophysical CCM in algal cultures to study its relative contribution to carbon fixation [2]. |
| 3-Mercaptopicolinic Acid (MPA) | An inhibitor of phosphoenolpyruvate carboxykinase (PEPCK). Used to suppress the biochemical CCM (C4-like pathway) in algae [2]. |
| Carbonic Anhydrase (Exogenous) | An enzyme added to microalgal cultures in photobioreactors to stimulate the biophysical CCM, enhancing HCO₃⻠transport and improving CO₂ fixation efficiency under high CO₂ conditions [41]. |
| Polygenic Priority Score (PoPS) | A computational method that integrates multi-dimensional genomic features to enhance the functional prediction and prioritization of candidate genes from GWAS hits, moving beyond simple nearest-gene annotation [39]. |
| Pterygospermin | Pterygospermin, CAS:11054-42-5, MF:C22H18N2O2S2, MW:406.5 g/mol |
| Pap12-6 | Pap12-6, MF:C83H133N23O12, MW:1645.1 g/mol |
Experimental Workflows & Pathway Diagrams
Workflow for Multi-Omics CCM Gene Discovery
Coordination Between CCMs in Algae
Inhibitor-Based Experimental Design
The quest to enhance carbon fixation is a central challenge in synthetic biology and climate change mitigation. In nature, many algae and cyanobacteria utilize specialized structures known as CO2-concentrating mechanisms (CCMs) to supercharge photosynthesis. Research on the green macroalga Ulva prolifera demonstrates a sophisticated complementary coordination between biophysical and biochemical CCMs. When its biophysical CCM was inhibited, the biochemical CCM compensated for approximately 50% of total carbon fixation. Conversely, the biophysical CCM could compensate for nearly 100% of fixation when the biochemical CCM was inhibited, showcasing a robust, flexible system for maintaining photosynthetic efficiency [2] [42].
Synthetic biology aims to transfer such efficient natural systems into industrially relevant organisms and crops. However, installing natural compartments like cyanobacterial carboxysomes into plants is challenging due to their genetic complexity and specificity. A groundbreaking alternative uses simpler bacterial protein cages called encapsulins from Quasibacillus thermotolerans (QtEnc) [43] [44]. These nanocompartments can be reprogrammed into modular carbon-fixing organelles, providing a simplified, isoform-agnostic platform for constructing synthetic CCMs. This technical support center details the methodologies and troubleshooting for implementing this technology within the broader context of optimizing CCM coordination.
Frequently Asked Questions (FAQs)
Q1: What are the primary advantages of using encapsulins over native carboxysomes for building synthetic CCMs? Encapsulins offer a modular and streamlined alternative to native carboxysomes. While carboxysomes require the precise expression and balance of multiple shell proteins and can only package their native Rubisco, the encapsulin system from Quasibacillus thermotolerans (QtEnc) is encoded by a single gene and self-assembles. Most importantly, it is isoform-agnostic; by fusing a short cargo-loading peptide (CLP) to diverse Rubisco isoforms, researchers can achieve targeted encapsulation without extensive genetic redesign [43] [44] [45].
Q2: How is Rubisco engineered for encapsulation, and does this affect its function? Rubisco is engineered by fusing a short cargo-loading peptide (CLP)—14 amino acids for QtEnc—to the enzyme. Structure-function considerations are critical:
- For Form I Rubiscos (e.g., from tobacco or R. sphaeroides), the CLP is typically appended via a flexible linker to the C-terminus of the RbcS small subunit. This location is structurally disordered and distant from the catalytic sites, minimizing functional perturbation [43].
- For Form II Rubisco (e.g., from R. rubrum), which lacks small subunits, the CLP can be fused to either the N- or C-terminus of the RbcL large subunit [43]. Studies confirm that with the correct tagging strategy, the oligomeric assembly of Rubisco is preserved, and it retains its CO2-fixing activity after encapsulation [43].
Q3: What is the role of carbonic anhydrase (CA) in a functional CCM, and is it part of the current encapsulin system? Carbonic anhydrase (CA) is a vital component of biophysical CCMs. It catalyzes the interconversion of bicarbonate (HCO3-) and CO2, ensuring a high local concentration of CO2 is supplied to Rubisco within the compartment [2] [41]. The current encapsulin-based system described in the foundational research is a proof-of-concept that successfully encapsulates active Rubisco. However, the authors note that carbonic anhydrase remains to be incorporated into the nanocompartment to create a fully functional, synthetic CCM [43] [46].
Q4: Our encapsulated Rubisco shows poor catalytic activity. What could be the cause? Poor activity can stem from several factors:
- Incorrect CLP Tagging Site: Fusing the CLP to a critical functional region of Rubisco (e.g., the C-terminus of RbcL in Form I Rubiscos) can interfere with the enzyme's "latching" mechanism and catalytic fidelity [43].
- Improper Assembly Order: For complex Rubisco isoforms, simultaneous co-expression of the enzyme and the encapsulin shell can lead to misassembly. A staged induction protocol—producing Rubisco first, then inducing the shell—is often necessary for correct assembly and function [44] [45].
- Missing CCM Components: The current activity assays measure Rubisco's inherent function. The full catalytic potential will only be realized once the CO2-concentrating machinery, including carbonic anhydrase and bicarbonate transporters, is integrated into the system [43].
Troubleshooting Guide
| Problem Area | Specific Issue | Possible Cause | Recommended Solution |
|---|---|---|---|
| Protein Expression & Assembly | Low yield of CLP-tagged Rubisco | Fusion tag disrupts folding/assembly; proteolytic degradation. | Optimize expression conditions (temp., inducer concentration); test solubility tags; use protease-deficient strains. |
| Incomplete nanocompartment formation | Imbalanced expression; improper self-assembly conditions. | Tune expression ratio of Rubisco:Encapsulin; use staged induction protocol [44]. | |
| Encapsulation Efficiency | Rubisco not loaded into encapsulin | CLP tag is inaccessible; shell pores are restrictive. | Verify tag placement on solvent-exposed, flexible termini (e.g., RbcS C-terminus) [43]; analyze shell mutants with larger pores. |
| Heterogeneous nanocompartment populations | Incomplete or malformed assembly. | Purify assemblies using sucrose density gradient centrifugation to isolate fully formed, Rubisco-loaded compartments. | |
| Enzymatic Function | Encapsulated Rubisco has low catalytic activity | CLP tag disrupts active site; crowded interior environment. | Re-engineer tag location (e.g., from RbcL to RbcS); ensure shell pores allow substrate/product diffusion [43] [44]. |
| High oxygenation vs. carboxylation | Lack of localized CO2 concentration. | Integrate carbonic anhydrase into the system as the next critical development step [43]. |
Key Experimental Protocols
Engineering and Expressing CLP-Tagged Rubisco
This protocol outlines the process for creating and producing Rubisco enzymes ready for encapsulation.
Methodology:
- Gene Construction: Fuse the DNA sequence encoding the 14-amino-acid QtEnc CLP to the gene of your target Rubisco. For Form I Rubiscos (L8S8), append the CLP via a 6-residue glycine-serine linker to the 3' end of the RbcS (small subunit) gene. For the Form II Rubisco (L2), test both N- and C-terminal fusions on the RbcL gene [43].
- Expression in E. coli: Clone the constructed gene into an appropriate expression vector. Transform into a suitable E. coli strain (e.g., BL21(DE3)).
- Induction and Analysis: Induce expression with IPTG. Confirm expression and oligomeric assembly using SDS-PAGE and native PAGE. The CLP-tagged Rubisco should show a clear shift in mobility on native gels compared to the untagged version, indicating proper assembly and increased molecular mass [43].
Assembling Rubisco-Filled Encapsulin Nanocompartments
This protocol describes two strategies for assembling the full nanocompartment.
Methodology: There are two primary methods for assembly:
- A. Co-induction: Co-express the CLP-tagged Rubisco and the QtEnc shell protein simultaneously from a single or dual plasmid system. This is simpler but may be less effective for complex Rubiscos [43].
- B. Staged Induction: This method is preferred for more complex Rubisco isoforms.
After cell lysis, the intact nanocompartments can be purified using methods like nickel-affinity chromatography (if his-tagged) and sucrose density gradient centrifugation, which effectively separates loaded from empty compartments [43].
Functional Analysis of Carbon Fixation
This protocol covers how to verify the functionality of the constructed nanocompartments.
Methodology:
- In vitro Activity Assay: Measure the carboxylase activity of the purified, encapsulated Rubisco. The assay typically uses radiolabeled 14CO2 or a coupled enzymatic system to track the incorporation of inorganic carbon into 3-phosphoglycerate. Compare the activity to an equivalent amount of free, untagged Rubisco to determine the effect of encapsulation [43].
- Validation in Model Organisms: Preliminary plant experiments are the next step. Introduce the genes for the CLP-tagged Rubisco and QtEnc shell into the model plant Nicotiana benthamiana or target crops like rice and wheat. Analyze the resulting plants for nanocompartment formation, Rubisco activity, and overall photosynthetic performance [44] [45].
The Scientist's Toolkit: Research Reagent Solutions
| Item Name | Function / Application | Key Details & Considerations |
|---|---|---|
| QtEnc (Quasibacillus thermotolerans Encapsulin) | Self-assembling protein nanocage scaffold. | Forms a 42 nm icosahedral compartment; requires only one gene for shell formation; pores allow substrate/product diffusion [43] [44]. |
| Cargo-Loading Peptide (CLP) | Directs specific cargo encapsulation. | A 14-amino-acid peptide tag; fused to cargo proteins like Rubisco; acts as a "postcode" for encapsulation [43] [45]. |
| Rubisco Isoforms (Nt, Rs, Rr) | Core carbon-fixing enzyme for encapsulation. | Test diverse isoforms (e.g., Tobacco-Nt, R. sphaeroides-Rs, R. rubrum-Rr); tagging strategy is isoform-dependent [43]. |
| Carbonic Anhydrase (CA) | Future component for a complete CCM. | Catalyzes HCO3- to CO2 conversion; essential for creating a high-CO2 microenvironment around Rubisco; not yet integrated in current system [43] [41]. |
| EZ (Ethoxyzolamide) & MPA (3-Mercaptopicolinic Acid) | Pharmacological inhibitors for CCM studies. | EZ inhibits CA (biophysical CCM); MPA inhibits PEPCK (biochemical CCM). Useful for probing CCM function and compensation in algal models [2] [42]. |
| Reveromycin B | Reveromycin B, MF:C36H52O11, MW:660.8 g/mol | Chemical Reagent |
The development of encapsulin-based carbon-fixing nanocompartments represents a pivotal step towards engineering synthetic CO2-concentrating mechanisms. The future research roadmap involves several critical steps to move from this powerful proof-of-concept to a fully functional system in plants. The immediate next step is the integration of carbonic anhydrase to create the necessary CO2-rich microenvironment for Rubisco [43] [41]. Subsequently, the entire system must be successfully expressed and assembled in chloroplasts, with the ultimate goal of transferring this technology to major C3 crops like wheat and rice to boost yields and resource-use efficiency [44] [45].
This approach, inspired by the coordinated CCMs found in algae, offers a more tractable and modular path to enhancing photosynthesis. By providing these detailed protocols, FAQs, and troubleshooting guides, this technical support center aims to empower researchers to adopt, optimize, and advance this promising technology, contributing to the broader goal of optimizing carbon fixation for food security and environmental sustainability.
Diagrams & Visualizations
Encapsulin Nanocompartment Workflow
CCM Coordination in Algal Research
Optimization Strategies and Challenge Mitigation in CCM Engineering
Troubleshooting Guides & FAQs
How can I determine if a biophysical CCM is malfunctioning in my algal cultures, and what are the key indicators?
A breakdown in the biophysical CO2 Concentrating Mechanism (CCM) primarily affects the active transport and conversion of inorganic carbon (Ci), leading to a direct drop in photosynthetic efficiency.
Key Indicators of Failure:
- Reduced Carbon Fixation: A measurable decline in photosynthetic O2 evolution or growth rate, especially when transitioning from high to low CO2 conditions [7] [2].
- Sensitivity to Low CO2: Impaired photoautotrophic growth under air-level (0.04% - L-CO2) or very low (0.01% - VL-CO2) CO2 conditions, while growth remains normal under high CO2 (2-5%) [7].
- Defective Enzyme Function: Inhibition of carbonic anhydrase (CA), a key enzyme in the biophysical CCM that catalyzes the interconversion of HCO3- and CO2, severely compromises carbon fixation [2].
Diagnostic Experimental Protocol:
- Objective: To assess the functional state of the biophysical CCM by measuring photosynthetic O2 evolution with and without a CA inhibitor.
- Materials:
- Algal culture (e.g., Ulva prolifera, Chlamydomonas reinhardtii)
- Clark-type O2 electrode system
- Buffered artificial seawater (for marine species) or medium (for freshwater species), pH 8.0
- Sodium bicarbonate (NaHCO3) stock solution
- Ethoxyzolamide (EZ), a potent inhibitor of both external and internal CA [2]
- Methodology:
- Harvest algal samples and acclimatize them in Ci-free buffered medium for 30 minutes to deplete internal Ci reserves [2].
- Resuspend the samples in fresh buffered medium containing 2 mM NaHCO3.
- Measure the baseline rate of photosynthetic O2 evolution.
- Add EZ to the medium at a final concentration of 50 µM and measure the O2 evolution rate again [2].
- Calculate the percentage inhibition of photosynthesis:
[1 - (Rate with EZ / Rate without EZ)] × 100.
- Interpretation: A significant inhibition (>50%) of photosynthetic O2 evolution after EZ addition indicates a heavy reliance on a functional biophysical CCM. A weak response suggests the CCM is not fully operational or that alternative pathways are compensating [2].
What are the experimental signs of a disrupted biochemical CCM, and how does it differ from a biophysical CCM failure?
A dysfunctional biochemical CCM, often involving a C4-like pathway, impairs the conversion of inorganic carbon into organic C4 acids before final fixation by Rubisco. This often manifests under specific environmental stresses.
Key Indicators of Failure:
- Accumulation of Photorespiratory Metabolites: Increased excretion of glycolate, a key photorespiratory intermediate, indicates a disruption in the photorespiratory cycle, which is closely linked to CCM function [7].
- Growth Defects Under Specific Stressors: The biochemical CCM may become critical under conditions like zinc deficiency, where the biophysical CCM is impaired [2].
- Inhibited C4 Acid Metabolism: Use of phosphoenolpyruvate carboxykinase (PEPCK) inhibitors, such as 3-mercaptopicolinic acid (MPA), specifically targets this biochemical pathway [2].
Diagnostic Experimental Protocol:
- Objective: To evaluate the contribution of the biochemical CCM by inhibiting PEPCK and measuring the impact on photosynthesis.
- Materials:
- Algal culture
- Clark-type O2 electrode system
- Buffered medium, pH 8.0
- NaHCO3 stock solution
- 3-mercaptopicolinic acid (MPA) stock solution [2]
- Methodology:
- Follow the same sample preparation and baseline measurement steps as in the biophysical CCM diagnostic protocol.
- Add MPA to the medium at a final concentration of 1.5 mM and measure the new O2 evolution rate [2].
- Calculate the percentage inhibition of photosynthesis.
- Interpretation: Significant inhibition by MPA points to a active biochemical CCM contributing to carbon fixation. Research on Ulva prolifera showed that when the biochemical CCM was inhibited, the biophysical CCM could compensate for almost 100% of carbon fixation, demonstrating a robust coordination mechanism [2].
What does a coordination breakdown between biophysical and biochemical CCMs look like, and what are its systemic consequences?
A coordination failure means the two systems do not complement each other effectively, leading to an overall reduction in metabolic plasticity and fitness. This is often observed in specific mutants or under abrupt environmental changes.
Key Indicators of Failure:
- Failed Acclimation to Low CO2: Mutants like lci20 in Chlamydomonas, which is defective in a chloroplast envelope glutamate/malate transporter, show severe growth impairment specifically during a sudden transition from high to very low CO2 conditions. This indicates a failure in the integrated metabolic network supporting CCM, which involves energy shuttling between organelles [7].
- Impaired Downstream Biosynthesis: The pyrenoid, a central organelle for the biophysical CCM, also coordinates with other carbon-utilizing pathways. Mutants with a disrupted pyrenoid show impaired fatty acid and triacylglycerol biosynthesis, indicating a breakdown in the channeling of fixed carbon to anabolic pathways [8].
- Concurrent Activation of Compensatory Pathways: When one CCM is inhibited, the other should be reinforced. For example, when the biochemical CCM in Ulva was inhibited, the biophysical CCM was observed to compensate. A lack of such compensation suggests poor coordination [2].
Systemic Consequences: A breakdown in CCM coordination not only reduces the efficiency of photosynthesis but also has broader implications:
- Disrupted Energy Trafficking: The malate shuttle, involving transporters like LCI20, is crucial for transferring energy (as reducing power) between the chloroplast and mitochondria to power CCM components. Its disruption causes energy imbalance [7].
- Photorespiratory Stress: When the CCM is not properly induced or coordinated, photorespiration remains active even under low CO2, potentially leading to the accumulation of toxic intermediates like 2-phosphoglycolate (2-PG) if not properly managed through excretion or recycling [7].
The diagram below illustrates the ideal coordination between biophysical and biochemical CCMs and the points where breakdowns commonly occur, leading to the symptoms described in the troubleshooting guides.
What quantitative data can I expect from inhibitor experiments studying CCM contributions?
The table below summarizes typical quantitative findings from experiments using specific inhibitors to dissect the contributions of biophysical and biochemical CCMs, as demonstrated in studies on algae like Ulva prolifera.
Table 1: Quantitative Contributions of CCMs to Photosynthetic Carbon Fixation
| Algal Species | Inhibitor Used | Target Pathway | Observed Inhibition of Carbon Fixation | Interpretation & Context |
|---|---|---|---|---|
| Ulva prolifera | Ethoxyzolamide (EZ) | Biophysical CCM (Carbonic Anhydrase) | ~50% decline [2] | Indicates a dominant role for the biophysical CCM under tested conditions. |
| Ulva prolifera | 3-mercaptopicolinic acid (MPA) | Biochemical CCM (PEPCK) | Carbon fixation declined; biophysical CCM compensated for ~100% of total fixation [2] | Biochemical CCM plays a supporting role; demonstrates high compensatory capacity of biophysical CCM. |
What essential reagents and materials are needed for troubleshooting CCM imbalances?
A well-stocked laboratory should have the following key reagents to effectively diagnose and research CCM imbalances in algal systems.
Table 2: Research Reagent Solutions for CCM Analysis
| Reagent / Material | Function & Application | Key Consideration |
|---|---|---|
| Ethoxyzolamide (EZ) | A potent inhibitor of carbonic anhydrase (CA). Used to suppress the biophysical CCM and evaluate its contribution to photosynthesis [2]. | Inhibits both external and intracellular CA. Use with appropriate controls. |
| 3-Mercaptopicolinic Acid (MPA) | A specific inhibitor of phosphoenolpyruvate carboxykinase (PEPCK). Used to inhibit the biochemical (C4-like) CCM [2]. | Validates the operation and contribution of a C4 metabolic pathway in algae. |
| Clark-type Oâ‚‚ Electrode | Core instrument for measuring the rate of photosynthetic oxygen evolution, a direct proxy for carbon fixation efficiency [2]. | Requires careful calibration. Samples must be depleted of internal Ci prior to assay for accurate results. |
| Mutant Strains (e.g., cia5, lci20) | Genetically modified algal lines (e.g., in Chlamydomonas reinhardtii) with specific defects in CCM genes. Essential for dissecting the genetic basis of CCM coordination [7]. | The cia5 mutant is defective in the master regulator of CCM induction. The lci20 mutant is defective in a chloroplast transporter linking metabolism. |
| LCI20 Antibody | A custom antibody used to detect the presence and localization of the LCI20 transporter protein via immunoblot analysis, confirming mutant phenotypes [7]. | Critical for validating genetic constructs and confirming protein-level expression in wild-type vs. mutant strains. |
Visualization of a Key Experimental Workflow
The following diagram outlines a standard experimental workflow for diagnosing CCM imbalances, integrating the reagents and methods discussed in the FAQs.
FAQs & Troubleshooting Guides
Frequently Asked Questions
1. What are the first signs that my algal culture is actively employing a CCM? The most direct initial sign is the transcriptional upregulation of key CCM and photorespiration genes, often controlled by the master regulator CIA5 (also called CCM1) [7] [47]. Physiologically, you may observe a downregulation of photoprotective proteins like LHCSR3 and PSBS, as an active CCM elevates internal COâ‚‚, suppressing the need for energy dissipation [47]. Under the microscope, re-localization of proteins like acetyl-CoA carboxylase subunits to the pyrenoid periphery can also indicate CCM activation [8].
2. I am not seeing the expected growth phenotype in my CCM mutant under low COâ‚‚. What could be wrong?
The growth phenotype can be condition-dependent. Some mutants, like the lci20 mutant defective in a chloroplast envelope malate/glutamate transporter, show a severe growth defect only during a sudden transition from high to very-low COâ‚‚ conditions, but grow normally if pre-acclimated [7]. Ensure your experimental protocol includes such transition stress tests. Furthermore, verify the Oâ‚‚ concentration, as a photorespiratory phenotype might be masked under non-photorespiratory conditions (e.g., 2% Oâ‚‚) [7].
3. How can I determine whether the biophysical or biochemical CCM is more active in my strain under a specific stress? The relative contribution of each CCM can be quantified using specific metabolic inhibitors in combination with measurements of photosynthetic carbon fixation or Oâ‚‚ evolution [2].
- To inhibit the biophysical CCM, use ethoxyzolamide (EZ), which targets carbonic anhydrase activity.
- To inhibit the biochemical CCM, use 3-mercaptopicolinic acid (MPA), which targets phosphoenolpyruvate carboxykinase (PEPCK). The drop in carbon fixation upon application of each inhibitor indicates the relative contribution of that CCM pathway. Research on Ulva prolifera showed the biophysical CCM can compensate for nearly 100% of fixation if the biochemical CCM is impaired, while the biochemical CCM contributes about 50% if the biophysical CCM is inhibited [2].
4. Why are my algae excreting glycolate, and is this a problem for my experiments? Glycolate excretion is a natural phenomenon in many algae, like Chlamydomonas, under conditions where photorespiration is active [7]. It serves as a safety valve to prevent the toxic accumulation of photorespiratory intermediates like 2-phosphoglycolate (2-PG) [7]. While it represents a loss of fixed carbon, it is not necessarily a problem for your culture but is a key metabolic indicator. It confirms that photorespiratory pressure exists, meaning the CCM is not fully suppressing Rubisco's oxygenase activity. Monitoring glycolate excretion can be a useful marker for the metabolic status of your cells.
Troubleshooting Common Experimental Issues
Table: Troubleshooting CCM-Related Experimental Problems
| Problem | Possible Cause | Solution |
|---|---|---|
| Poor growth under low COâ‚‚, but CCM genes are induced. | Energy (ATP) limitation for bicarbonate transporters [7] [48]. | Ensure adequate light quality and intensity. Investigate potential disruption to mitochondrial metabolism and associated malate shuttles that supply ATP [7]. |
| Unexpectedly low LHCSR3 protein levels under high light. | Repression by high intracellular COâ‚‚ from acetate metabolism in the media or high external COâ‚‚ [47]. | Omit acetate from the growth medium for photoprotection studies and rigorously control COâ‚‚ bubbling levels. Use metabolic mutants (e.g., icl) to dissect effects [47]. |
| Inconsistent CCM induction data between replicates. | Incomplete acclimation to new COâ‚‚ conditions; fluctuating dissolved COâ‚‚ in the medium. | Standardize and extend the acclimation period after changing COâ‚‚ levels. Use pH-stat systems or calibrated gas mixing systems to maintain a stable and precise dissolved COâ‚‚ concentration [48]. |
| No phenotype in a putative CCM mutant. | Genetic redundancy (e.g., multiple bicarbonate transporters) or compensatory activation of alternative CCM pathways [2]. | Perform a double or triple mutant analysis. Use inhibitor studies (e.g., EZ, MPA) on the mutant to uncover compensatory mechanisms [2]. |
Detailed Experimental Protocols
Protocol 1: Differentiating Biophysical vs. Biochemical CCM Contribution
This protocol uses specific metabolic inhibitors to quantify the relative contribution of each CCM to total photosynthetic carbon fixation [2].
Key Reagents:
- Ethoxyzolamide (EZ): A permeant inhibitor of carbonic anhydrase, used to suppress the biophysical CCM.
- 3-Mercaptopicolinic Acid (MPA): An inhibitor of phosphoenolpyruvate carboxykinase (PEPCK), used to suppress the biochemical CCM.
- Buffered artificial seawater or culture medium (pH 8.0).
- NaH¹â´CO₃ or Clark-type Oâ‚‚ electrode system.
Methodology:
- Culture Preparation: Grow algal cells to mid-log phase under the desired stress condition (e.g., low COâ‚‚, high light, zinc limitation).
- Ci Depletion: Harvest and transfer cells to Ci-free buffered medium. Gently aerate with Nâ‚‚ for 30 minutes to deplete endogenous inorganic carbon sources.
- Inhibitor Incubation: Divide the culture into four aliquots:
- Control: No inhibitor.
- +EZ: Add EZ to a final concentration of 50 µM.
- +MPA: Add MPA to a final concentration of 1.5 mM.
- +EZ+MPA: Both inhibitors.
- Incubate for 15-30 minutes.
- Carbon Fixation Assay: Add 2 mM NaH¹â´CO₃ to each aliquot. For radioactive tracing, measure incorporated ¹â´C after a fixed time (e.g., 10-30 min). Alternatively, use a Clark-type Oâ‚‚ electrode to measure rates of photosynthetic Oâ‚‚ evolution.
- Data Analysis:
- The inhibition in the +EZ treatment indicates the contribution of the biophysical CCM.
- The inhibition in the +MPA treatment indicates the contribution of the biochemical CCM.
- The response in the +EZ+MPA treatment reveals any non-CCM-related compensatory pathways.
Protocol 2: Monitoring Metabolic Shifts via Glycolate Excretion
This protocol assesses photorespiratory activity and the "overflow" capacity of the algae under CCM stress [7].
Key Reagents:
- Specific assay kit for glycolate (enzymatic or HPLC-based).
- Culture medium without organic carbon sources.
Methodology:
- Stress Application: Subject algal cultures to the stressor of interest (e.g., shift from high to very-low COâ‚‚).
- Conditioned Medium Collection: At defined time points (e.g., 0, 6, 12, 24h post-stress), take culture samples and centrifuge to pellet cells. Collect the cell-free supernatant.
- Glycolate Quantification: Analyze the supernatant for glycolate concentration using a commercial enzymatic assay kit or via HPLC.
- Data Interpretation: An increase in glycolate excretion indicates active photorespiration and potential saturation or incomplete induction of the CCM. This is often coordinated with the down-regulation of glycolate dehydrogenase [7].
The Scientist's Toolkit: Key Research Reagents
Table: Essential Reagents for Studying Algal CCM Compensation
| Reagent | Function / Target | Example Use in CCM Research |
|---|---|---|
| CIA5/CCM1 Mutants | Master transcriptional regulator of CCM and photorespiratory genes [7] [47]. | Serves as a positive control for CCM-deficient phenotypes; used to dissect the regulatory hierarchy. |
| EZ (Ethoxyzolamide) | Inhibitor of carbonic anhydrase (CA) [2]. | Suppresses the biophysical CCM by blocking HCO₃â»/COâ‚‚ interconversion, allowing quantification of its contribution. |
| MPA (3-Mercaptopicolinic Acid) | Inhibitor of PEPCK, a key decarboxylase in algal C4-like biochemistry [2]. | Suppresses the biochemical CCM to evaluate its role and the compensatory capacity of the biophysical CCM. |
lci20 Mutant |
Defective in a chloroplast envelope glutamate/malate transporter [7]. | Used to study the integration of photorespiration with CCM, and the role of metabolite shuttles in energy supply for CCM. |
icl Mutant |
Defective in isocitrate lyase, a key enzyme of the glyoxylate cycle [47]. | Used to dissect the source of COâ‚‚ from organic carbon metabolism (e.g., acetate) and its signaling effect on CCM/photoprotection genes. |
| Pyrenoid-Deficient Mutants | e.g., mutants with disrupted pyrenoid structure or Rubisco packaging. | Used to study the link between pyrenoid integrity and downstream processes like fatty acid biosynthesis [8]. |
Visualizing CCM Coordination and Regulation
The following diagram illustrates the core regulatory logic and compensatory interactions between the biophysical and biochemical CCMs in response to low COâ‚‚ stress, based on recent findings.
Diagram: COâ‚‚ Sensing and CCM Coordination Logic. This map outlines the regulatory network through which algae sense low COâ‚‚ and activate compensatory CCM pathways. The master regulator CIA5 is activated under low COâ‚‚ stress, simultaneously inducing both the Biophysical CCM (primary) and Biochemical CCM (secondary) [7] [47]. Successful CCM operation elevates internal COâ‚‚, which subsequently represses photoprotective mechanisms [47]. A key compensatory loop exists: impairment of one CCM type triggers the boosting of the other [2]. The pyrenoid, a central organelle for the biophysical CCM, also plays a role in supplying carbon to other pathways, such as fatty acid biosynthesis via acetyl-CoA carboxylase (ACC) condensates [8].
Frequently Asked Questions (FAQs)
FAQ 1: What are the most common metabolic bottlenecks that limit carbon fixation efficiency in microalgae?
The most common bottlenecks are found in the core pathways of central carbon metabolism. A significant constraint is the inherent inefficiency of the key enzyme RuBisCO, which has a low affinity for CO2 and is prone to initiating energy-wasting photorespiration [49]. Furthermore, limitations in the supply of essential precursors like acetyl-CoA and erythrose-4-phosphate (E4P) can restrict the synthesis of target products such as lipids and aromatic compounds [50]. Insufficient energy and redox cofactors (e.g., NADPH) also create bottlenecks, as they are required to drive biosynthetic reactions [49] [50].
FAQ 2: How can we experimentally distinguish between the contributions of biophysical and biochemical carbon concentration mechanisms (CCMs) in algal species?
You can distinguish their contributions using specific enzyme inhibitors in culture experiments [2].
- Inhibiting the Biophysical CCM: Use ethoxyzolamide (EZ), an inhibitor of carbonic anhydrase (CA). This enzyme is crucial for converting bicarbonate to CO2, and its inhibition directly impairs the biophysical CCM [2].
- Inhibiting the Biochemical CCM: Use 3-mercaptopicolinic acid (MPA), an inhibitor of phosphoenolpyruvate carboxykinase (PEPCK). This enzyme is a key component of the C4-biochemical CCM in some algae [2]. By measuring changes in photosynthetic carbon fixation rates (e.g., via O2 evolution) in the presence of these inhibitors, you can quantify the relative contribution of each CCM. Research on Ulva prolifera has shown that when one CCM is inhibited, the other can be reinforced to compensate, demonstrating a complementary coordination mechanism [2].
FAQ 3: What cultivation strategies can help overcome resource consumption bottlenecks in large-scale microalgae cultivation?
Two key strategies can significantly reduce resource consumption and cost [51]:
- Using Wastewater as a Culture Medium: This approach avoids the consumption of fresh water and provides a source of inorganic and organic nutrients (e.g., nitrogen, phosphorus). It integrates bioremediation with biomass production, improving sustainability [51].
- Adopting Mixed-Species Cultivation: Instead of maintaining fragile monocultures, cultivating a selected community of microalgae and bacteria can improve the stability of the system against contamination by pests and pathogens. This mixed community can be more resilient and productive when using wastewater [51].
Troubleshooting Guides
Table 1: Common Experimental Problems & Solutions
| Problem | Possible Cause | Suggested Solution |
|---|---|---|
| Low biomass productivity in photoautotrophic cultivation | Light saturation or photoinhibition; Low CO2 availability [51] [49] | Implement vertical mixing in the reactor; Supplement with CO2 (e.g., 2-5%) but avoid levels that cause acid stress [51] [49]. |
| Low yield of target products (e.g., lipids, carotenoids) | Metabolic bottlenecks in precursor supply (e.g., acetyl-CoA); Insufficient reducing power (NADPH) [49] [50] | Engineer heterologous pathways like phosphoketolase (PK) to enhance acetyl-CoA flux; Overexpress NADPH-generating enzymes in the PPP [50]. |
| Culture collapse in open ponds | Contamination by undesirable pests or pathogens [51] | Use selected wild, robust algal strains and shift to a mixed-species cultivation system to improve ecological stability [51]. |
| Inefficient harvesting of algal cells | Low cell density and similarity of cell density to water [51] | Explore new strategies such as attaching or absorbing cells onto solid materials to facilitate separation, moving beyond traditional centrifugation and filtration [51]. |
Table 2: Analysis of CCM Inhibitor Effects
| Inhibitor | Target Enzyme | CCM Type Affected | Expected Outcome on Carbon Fixation |
|---|---|---|---|
| Ethoxyzolamide (EZ) | Carbonic Anhydrase (CA) | Biophysical [2] | Significant decline in fixation rate, demonstrating this mechanism's dominant role. |
| 3-Mercaptopicolinic Acid (MPA) | Phosphoenolpyruvate Carboxykinase (PEPCK) | Biochemical (C4-like) [2] | Reduced fixation, indicating the supporting role of the biochemical CCM. |
Experimental Protocols
Protocol 1: Assessing CCM Contributions Using Inhibitors
This protocol allows for the quantitative assessment of the roles of biophysical and biochemical Carbon Concentration Mechanisms (CCMs) in algal photosynthesis.
Key Materials:
- Algal culture (e.g., Ulva prolifera)
- Specific inhibitors: Ethoxyzolamide (EZ) and 3-mercaptopicolinic acid (MPA)
- Clark-type O2 electrode system
- Buffered artificial seawater (pH 8.0)
Methodology:
- Culture Pre-treatment: Cut algal samples into fragments and acclimate them in Ci-free buffered artificial seawater for 30 minutes to deplete internal carbon stores [2].
- Inhibitor Preparation: Prepare experimental solutions with 2 mmol/L NaHCO3 and add inhibitors to final concentrations of 50 µmol/L for EZ and 1.5 mmol/L for MPA [2].
- O2 Evolution Measurement: Place the pre-treated algal fragments into the O2 electrode chamber containing the buffered seawater with NaHCO3 and the chosen inhibitor.
- Data Acquisition: Measure the rate of photosynthetic O2 evolution under constant light and temperature (e.g., 200 µmol photons mâ»Â² sâ»Â¹ and 22°C).
- Analysis: Compare the O2 evolution rates with and without inhibitors. The percentage inhibition is calculated as:
[1 - (Rate with inhibitor / Rate without inhibitor)] * 100[2].
The workflow for this experiment is outlined below:
Protocol 2: Enhancing Carbon Channeling via the PHK Pathway
This genetic engineering protocol aims to re-route central carbon metabolism to alleviate bottlenecks in the production of acetyl-CoA-derived compounds.
Key Materials:
- S. cerevisiae or microalgal chassis strain
- Vectors for expressing phosphoketolase (PK) and phosphotransacetylase (PTA) genes
- Low-cost carbon source (e.g., glucose)
Methodology:
- Strain Engineering: Introduce heterologous genes encoding for the phosphoketolase (PK) and phosphotransacetylase (PTA) enzymes into the host chassis [50].
- Cultivation: Grow the engineered strain in a suitable medium with a carbon source like glucose.
- Pathway Activation: The PHK pathway directly converts fructose-6-phosphate (F6P) and xylulose-5-phosphate (X5P) from glycolysis and the PPP into acetyl-CoA, bypassing several steps in the central metabolism [50].
- Outcome: This rerouting increases the flux of carbon toward acetyl-CoA, a key precursor for lipids and other products, and can help correct redox imbalances by providing a route for NADPH consumption [50].
The metabolic rerouting effected by this protocol is shown in the following diagram:
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for CCM and Metabolic Bottleneck Research
| Reagent | Function / Target | Brief Explanation |
|---|---|---|
| Ethoxyzolamide (EZ) | Carbonic Anhydrase (CA) Inhibitor [2] | Used to inhibit the biophysical CCM by blocking the interconversion of bicarbonate and CO2, both extracellularly and intracellularly. |
| 3-Mercaptopicolinic Acid (MPA) | PEP Carboxykinase (PEPCK) Inhibitor [2] | Used to inhibit the biochemical CCM in organisms with C4-like metabolism by blocking the decarboxylation of C4 acids. |
| Phosphoketolase (PK) Gene | Heterologous Metabolic Enzyme [50] | Introduced to create a synthetic PHK pathway, directly converting sugar phosphates into acetyl-phosphate, thereby bypassing native glycolytic bottlenecks. |
| ATP:citrate lyase (ACL) Gene | Heterologous Metabolic Enzyme [50] | Provides an alternative route for cytosolic acetyl-CoA production directly from citrate, linking the TCA cycle to biosynthetic pathways. |
| Wastewater Medium | Low-Cost Nutrient Source [51] | Replaces freshwater and synthetic nutrients in cultivation, providing essential macro/micronutrients while reducing production costs and performing bioremediation. |
Conceptual FAQs: Understanding Energy Partitioning in Algal CCMs
FAQ 1: What is the fundamental energy conflict between the CCM and carbon fixation?
The CO2-concentrating mechanism (CCM) and the Calvin-Benson-Bassham (CBB) cycle are both major consumers of energy derived from the photosynthetic light reactions. The CCM actively consumes ATP to pump inorganic carbon (Ci) into the cell and chloroplast, concentrating CO2 around Rubisco. Simultaneously, the CBB cycle requires both ATP and NADPH to fix carbon and produce sugars. This creates a direct competition for a finite pool of cellular energy, particularly under limiting light or CO2 conditions. Efficient partitioning of energy between these two processes is therefore critical for maximizing photosynthetic growth [13].
FAQ 2: How does mitochondrial respiration interact with the chloroplast CCM?
Mitochondria play a surprisingly active role in supporting the CCM, especially under low CO2 conditions. Research on Chlamydomonas reinhardtii has shown that upon acclimation to low CO2, mitochondria migrate to a position between the chloroplast envelope and the plasma membrane. This strategic positioning suggests a role in supplying energy, potentially in the form of ATP produced by respiration, to power the CCM. Furthermore, mutations affecting mitochondrial complex I can alter chloroplast electron transport, influencing the redox state of the plastoquinone pool and thereby affecting photosynthetic energy generation available for CCM processes [52] [13].
FAQ 3: What is the role of proton gradients in CCM function?
Proton gradients are a central form of energy currency for the CCM. The active transport of bicarbonate (HCO3-) across biological membranes often relies on the proton motive force. For instance, some HCO3- transporters are coupled to H+ antiport. The acidification of specific compartments, driven by proton pumps, can also facilitate the conversion of HCO3- to CO2, making it available for Rubisco. The thylakoid membrane generates a large proton gradient (ΔpH) during photosynthesis for ATP synthesis, and a similar principle, though with different components, applies to Ci transport across the plasma membrane and chloroplast envelopes [53] [13].
FAQ 4: How do we quantitatively measure the ATP/NADPH demand of the CCM versus biomass production?
Stoichiometric metabolic modeling, such as Flux Balance Analysis (FBA), is a key tool for evaluating these demands. A recent network-wide analysis of alkene-producing Synechocystis revealed that biomass accumulation requires an ATP/NADPH ratio of 2.11 under autotrophic conditions, while the production of various alkenes required ratios below this value. This analysis calculates the turnover rates (flux-sums) of ATP and NADPH, providing a quantitative picture of cofactor usage and highlighting that the rate of NADPH regeneration is a key control point for the cellular ATP/NADPH ratio and bioproduction efficiency [54].
Table 1: Calculated Cofactor Turnover and Ratios for Biomass and Alkene Production in Synechocystis
| Objective | Growth Condition | ATP Turnover (mmol/gDW/h) | NADPH Turnover (mmol/gDW/h) | ATP/NADPH Ratio |
|---|---|---|---|---|
| Biomass | Autotrophic | 7.24 - 8.61 | 3.87 - 5.49 | 2.11 |
| Biomass | Mixotrophic | 7.24 - 8.61 | 3.87 - 5.49 | 19.49 |
| Alkene Production | Autotrophic | Varied | Varied | Below biomass ratio |
Source: Adapted from [54]
Troubleshooting Guides
Issue 1: Poor Growth and Photosynthetic Efficiency in Low CO2 Conditions
Problem: Your algal strain shows stunted growth or chlorosis (yellowing) when shifted from high CO2 (e.g., 2-5%) to ambient air or low CO2 conditions, indicating a failure to properly acclimate the CCM.
Investigation and Solution Path:
Confirm CCM Induction:
- Action: Check the expression levels of key CCM components. For Chlamydomonas reinhardtii, this includes genes like
HLA3andLCIAwhich are major Ci transporters, and carbonic anhydrases associated with the pyrenoid. - Tool: Use quantitative RT-PCR or available antibodies to verify that these genes are upregulated upon low-CO2 exposure.
- Rationale: Impaired CCM function can often be traced to faulty gene expression regulation rather than the primary energy supply [13].
- Action: Check the expression levels of key CCM components. For Chlamydomonas reinhardtii, this includes genes like
Diagnose Energy Supply to the CCM:
- Action: Assess the activity of cyclic electron flow (CEF) pathways.
- Protocol: Measure chlorophyll fluorescence parameters (e.g., NPQ, qE) which are dependent on the proton gradient (ΔpH) generated in part by CEF. Compare the kinetics of induction and relaxation between your strain and a wild-type control. A slow or low-magnitude NPQ response can indicate impaired CEF.
- Rationale: The PGR5-PGRL1 mediated CEF pathway is crucial for generating extra ATP without producing NADPH, which is thought to be important for powering the CCM [55] [13].
- Action: Evaluate mitochondrial function and its interaction with the chloroplast.
- Protocol: Use respiratory inhibitors (e.g., rotenone for Complex I) and measure the subsequent impact on photosynthetic O2 evolution or Ci affinity. Altered respiration can indirectly affect chloroplast energy status and CCM performance [52].
Issue 2: Imbalanced Cofactor Economy Leading to Metabolic Bottlenecks
Problem: Your engineered strain overproduces a target compound (e.g., isoprene) but exhibits much lower than predicted yields, or growth is severely penalized.
Investigation and Solution Path:
Quantify Cofactor Demand:
- Action: Perform a stoichiometric analysis of your engineered pathway.
- Tool: Use genome-scale metabolic models, if available for your strain, to calculate the theoretical ATP and NADPH demands of your product pathway versus biomass synthesis.
- Rationale: This helps identify if there is a cofactor imbalance. For example, a pathway that consumes too much NADPH may starve the CBB cycle, while an ATP-heavy pathway may compete directly with the CCM [54].
Engineer Cofactor Regeneration:
- Action: If an imbalance is identified, implement strategies to rebalance the cofactor pool.
- Solution A (High NADPH demand): Introduce non-native NADH-dependent oxidoreductases or engineer existing enzymes to use NADH instead of NADPH, freeing up NADPH for your pathway and the CBB cycle [54].
- Solution B (High ATP demand): Introduce an "ATP-wasting" cycle (e.g., expression of a cytosolic ATP hydrolase) to lower the ATP pool and stimulate its regeneration, which may enhance overall flux [54].
Issue 3: Inefficient Ci Uptake and Assimilation
Problem: Measurements show a low affinity for external Ci, suggesting a problem with the initial steps of the CCM.
Investigation and Solution Path:
Test Different Ci Forms:
- Action: Characterize photosynthesis using CO2 gas versus HCO3- dissolved in the medium.
- Protocol: Measure O2 evolution or chlorophyll fluorescence while sequentially adding known quantities of NaHCO3. Compare the affinity (Km) for Ci with a wild-type strain. A significantly higher Km points to a defect in HCO3- transport.
- Rationale: This can help isolate whether the problem is with CO2 or HCO3- uptake systems [56] [13].
Investigate the Pyrenoid Microenvironment:
- Action: Check the integrity and composition of the pyrenoid.
- Protocol: Use immunofluorescence or GFP-tagging to confirm the proper localization of Rubisco and essential pyrenoid components like the starch sheath or linker proteins (e.g., EPYC1 in Chlamydomonas).
- Rationale: A disorganized pyrenoid can lead to CO2 leakage, forcing the cell to expend more energy to maintain the internal CO2 concentration [56].
Key Experimental Protocols & Workflows
Protocol 1: Real-Time Monitoring of Stromal pH in Isolated Chloroplasts
The stromal pH is a key indicator of chloroplast energy status, alkalizing in the light to activate CBB cycle enzymes. This protocol uses the fluorescent dye BCECF-AM for non-destructive monitoring [57].
Workflow:
- Chloroplast Isolation: Isolate intact chloroplasts from plant tissue (e.g., pea shoots) via Percoll gradient centrifugation in grinding buffer (330 mM sorbitol, 50 mM HEPES-KOH pH 7.3) [57].
- Dye Loading: Incubate chloroplasts (0.5 mg/mL chlorophyll) with 20 µM BCECF-AM for 20 min at room temperature, then 10 min on ice [57].
- Remove External Dye: Re-isolate the chloroplasts through a 40% Percoll cushion and wash with grinding buffer [57].
- Fluorescence Measurement: Suspend the stained chloroplasts in a stirred cuvette in a fluorescence spectrometer. The ratio of fluorescence at excitation wavelengths 440 nm and 495 nm (emission at 535 nm) is ratiometrically correlated to the stromal pH [57].
- Calibration: Create a standard curve by measuring the fluorescence ratio at different known pH values (using buffers with ionophores to equilibrate internal and external pH) [57].
- Experimental Measurement: Illuminate the chloroplasts with actinic light. The established proton gradient across the envelope (ΔpHenv) can be calculated as [Measured Stromal pH] - [External Buffer pH] [57].
Protocol 2: Assessing the Energy State via Chlorophyll Fluorescence
Chlorophyll fluorescence measurements provide a non-invasive window into the photosynthetic light reactions and the proton gradient driving ATP synthesis.
Workflow:
- Dark Adaptation: Adapt the algal culture to darkness for at least 15 minutes.
- Measure Minimum Fluorescence (Fâ‚€): Apply a weak measuring light to get the baseline fluorescence.
- Measure Maximum Fluorescence (Fm): Apply a saturating pulse of light to close all PSII reaction centers.
- Calculate Dark-Adapted Parameters: Determine Fv/Fm = (Fm - Fâ‚€)/Fm as a measure of maximum PSII quantum yield.
- Actinic Illumination: Expose the sample to actinic light to drive photosynthesis.
- Measure Light-Adapted Parameters: Under steady-state light, measure F (steady-state fluorescence) and Fm' (maximum fluorescence under light).
- Induce and Relax NPQ: The rapid, energy-dependent quenching (qE) of NPQ is a direct reporter of thylakoid lumen acidification. Its induction and relaxation kinetics can report on the status of cyclic electron flow, which is critical for CCM energy supply [53] [55].
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Reagents for Investigating Energy Partitioning in Algal CCMs
| Reagent / Tool | Function / Target | Application in CCM Research |
|---|---|---|
| BCECF-AM [57] | Fluorescent pH probe (pKa ~6.98) | Non-destructive, real-time monitoring of stromal pH in isolated chloroplasts. |
| Nigericin [57] | K+/H+ ionophore | Collapses the H+ gradient (ΔpH) across chloroplast membranes to test its necessity for a process. |
| Rotenone [52] | Mitochondrial Complex I inhibitor | Used to dissect the role of mitochondrial respiration in supporting CCM function and energy metabolism. |
| Salicylhydroxamic Acid (SHAM) [52] | Alternative Oxidase (AOX) inhibitor | Inhibits the mitochondrial alternative respiratory pathway to assess its contribution to redox balancing under CCM-active conditions. |
| Genome-Scale Metabolic Model [54] | In silico stoichiometric network of metabolism | Predicts metabolic fluxes, identifies cofactor (ATP/NADPH) demands, and simulates outcomes of genetic manipulations. |
| Stoichiometric Flux Analysis (FBA) [54] | Computational constraint-based modeling | Calculates intracellular flux distributions, including ATP/NADPH turnover rates, to evaluate energy and redox balance during product synthesis. |
Frequently Asked Questions (FAQs)
Q1: We induced a CCM in our Chlamydomonas culture by shifting to low COâ‚‚ conditions, but observe reduced growth and glycolate excretion. Is the CCM functioning correctly, and why is photorespiration still active?
A1: Your observations are consistent with recent findings that the COâ‚‚ concentrating mechanism (CCM) and photorespiration can operate jointly in low COâ‚‚ environments, challenging the long-held belief that an active CCM fully suppresses photorespiration [58] [59]. Glycolate excretion acts as a safety valve to prevent the toxic accumulation of photorespiratory metabolites like 2-phosphoglycolate (2-PG) [58]. To troubleshoot:
- Verify that your CCM induction was successful by checking the expression of key CCM genes (e.g.,
HLA3for bicarbonate transport) and measuring internal inorganic carbon accumulation. - Monitor glycolate levels in the medium; significant excretion confirms active photorespiratory flux.
- Ensure your growth conditions (light intensity, Oâ‚‚ level) are not excessively promoting Rubisco oxygenase activity. Consider conducting experiments at 2% Oâ‚‚ to suppress photorespiration as a control [58].
Q2: Our mutant strain, defective in a key photorespiratory gene, shows severe growth impairment in air but not in high COâ‚‚. What is the underlying cause, and how can we investigate it?
A2: This is a classic phenotype indicating that the mutation affects a component essential for managing the 2-PG load under ambient, oxygen-containing conditions [60]. In high COâ‚‚, Rubisco's carboxylase activity is favored, minimizing 2-PG production and negating the need for the photorespiratory pathway. To investigate further:
- Confirm the metabolic block: Use metabolomics to profile and quantify photorespiratory intermediates (e.g., glycolate, glyoxylate). Accumulation of a specific metabolite can pinpoint the blocked reaction step [58].
- Check for redox imbalance: Photorespiration is linked to energy and redox metabolism. Measure ROS levels and the redox state of NAD(P)H pools, as disruptions can cause oxidative damage [61].
- Genetic complementation: Express the wild-type gene in your mutant to confirm that restoring the gene function rescues the normal growth phenotype in air [58].
Q3: We are attempting to reduce photorespiration in a microalgal strain by overexpressing CCM components. What is a critical, often-overlooked factor we should consider?
A3: A critical factor is the energy supply and trafficking between organelles. Active CCMs, especially those involving bicarbonate transporters like HLA3, require significant ATP [58] [59]. Overexpressing transporters without ensuring an adequate energy supply can lead to secondary metabolic stresses. Furthermore, recent research highlights the importance of metabolite shuttles (e.g., malate shuttles) involving chloroplast envelope transporters (e.g., LCI20) for providing energy and carbon skeletons from mitochondria to power the CCM [58] [59]. Ensure your engineering strategy includes enhancing the cell's energy generation and transport capacity.
Troubleshooting Guides
Problem: Poor Growth After Transition to Low COâ‚‚
| Observation | Potential Cause | Diagnostic Experiments | Solution |
|---|---|---|---|
| Acute growth impairment only during shift from high to very low COâ‚‚ [58] | Defect in chloroplast metabolite transporters (e.g., LCI20) required for photorespiration and energy supply [58]. | 1. Perform RT-qPCR to check LCI20 expression.2. Analyze metabolite profiles for accumulation of photorespiratory intermediates.3. Localize the protein via fluorescence tagging (e.g., LCI20-mVenus) to confirm chloroplast envelope presence [58]. |
Express the functional transporter gene in the mutant strain [58]. |
| Chronic poor growth under all low COâ‚‚ conditions [58] | Broader defect in CCM induction or function (e.g., cia5 mutation, pyrenoid defects). |
1. Check expression of core CCM genes (HLA3, CAH3).2. Immunofluorescence to inspect pyrenoid structure and Rubisco localization [58]. |
Ensure the master regulator CIA5 is functional; complement with missing CCM genes. |
Problem: Unexpected Glycolate Excretion
| Observation | Potential Cause | Diagnostic Experiments | Solution |
|---|---|---|---|
| High glycolate excretion under low COâ‚‚ with active CCM [58] | Native down-regulation of glycolate dehydrogenase (GlcDH) and active photorespiration; excretion is a detoxification mechanism [58]. | 1. Measure glycolate dehydrogenase enzyme activity.2. Quantify excreted glycolate in the culture medium. | This may be a normal physiological response. To reduce excretion, consider engineering a synthetic photorespiratory bypass pathway to fully metabolize glycolate. |
Glycolate excretion in a CCM-deficient mutant (e.g., cia5) at low COâ‚‚ [58] |
Severe photorespiration due to high Rubisco oxygenase activity without a COâ‚‚-concentrating mechanism. | 1. Genotype to confirm CCM deficiency.2. Grow cells under high COâ‚‚ or 2% Oâ‚‚; if excretion ceases, it confirms photorespiration is the source. | The CCM must be restored or the strain must be grown under high COâ‚‚ conditions to suppress photorespiration. |
Key Experimental Protocols
Protocol: Assessing CCM and Photorespiration Activity
Objective: To quantitatively evaluate the operational state of the COâ‚‚ concentrating mechanism and the photorespiratory pathway in algal cultures under different conditions.
Workflow Summary: The following diagram outlines the key decision points and analyses in this protocol.
Materials:
- Algal strain of interest (e.g., Chlamydomonas reinhardtii)
- Tris-Acetate-Phosphate (TAP) or minimal medium
- Controlled environment chambers (for COâ‚‚, Oâ‚‚, and light)
- Centrifuges
- RNA extraction kit
- RT-qPCR system
- HPLC system for organic acid analysis
- Equipment for targeted metabolomics (e.g., GC-MS, LC-MS)
Procedure:
- Culture Acclimation: Grow triplicate cultures of your algal strain under the conditions of interest (e.g., high COâ‚‚: 2-5%; low COâ‚‚: air level, ~0.04%; very low COâ‚‚: ~0.01%) for at least 48 hours to ensure full acclimation [58].
- Growth Monitoring: Measure growth rates daily by optical density (OD₆₈₀) and/or cell counting.
- Sample Collection:
- For glycolate excretion: Collect culture medium by centrifuging cells (e.g., 3,000 × g, 5 min). Filter the supernatant through a 0.22 μm filter and store at -20°C until analysis [58].
- For molecular analysis: Harvest cell pellets by centrifugation. Snap-freeze in liquid nitrogen and store at -80°C for RNA and metabolite extraction.
- Glycolate Quantification: Analyze the culture supernatant using HPLC to quantify excreted glycolate [58].
- Gene Expression Analysis: Extract total RNA from cell pellets. Perform RT-qPCR to measure transcript levels of key CCM genes (e.g.,
LCI20,HLA3,CAH3) and photorespiratory genes (e.g.,PGP1, GlcDH). Use housekeeping genes (e.g.,CBLP) for normalization [58]. - Metabolite Profiling: Perform targeted metabolomics on cell extracts to quantify key photorespiratory intermediates like 2-phosphoglycolate (2-PG), glycolate, and glycine [58].
Protocol: Characterizing a Putative Chloroplast Metabolite Transporter
Objective: To determine the function and localization of a putative chloroplast envelope transporter (e.g., LCI20) suspected to be involved in CCM/photorespiration coordination.
Workflow Summary: The diagram below illustrates the multi-faceted approach to characterize a putative transporter.
Materials:
- Putative transporter gene sequence (e.g.,
LCI20) - Corresponding mutant from the Chlamydomonas Library Project (CLiP) [58]
- Plasmid for genomic complementation (e.g., with PSAD promoter and mVenus tag) [58]
- Glass beads or electroporator for transformation
- Confocal microscope
- Antibody against the transporter (optional, for Western blot) [58]
Procedure:
- Obtain Mutant: Acquire the insertion mutant from the CLiP library. Verify the insertion and the absence of the full-length transcript via PCR and RT-PCR [58].
- Genetic Complementation: Clone the full-length genomic DNA of the putative transporter, including its promoter or under the control of a strong constitutive promoter (e.g., PSAD), into an expression vector. Fuse with a fluorescent protein tag (e.g., mVenus) for localization. Transform this construct into the mutant background [58].
- Protein Localization: Visualize the fluorescence in the complemented strain using confocal microscopy. Co-localization with chlorophyll fluorescence (chloroplast) and distinct envelope staining confirms a chloroplast envelope localization [58].
- Phenotypic Growth Assays: Spot the wild-type, mutant, and complemented strains on solid media (or monitor in liquid culture) under a matrix of conditions: High COâ‚‚, Low COâ‚‚, and Very Low COâ‚‚, combined with high (21%) and low (2%) Oâ‚‚. Document growth over 5-7 days [58]. A specific growth defect during the transition to very low COâ‚‚ is indicative of a role in acclimation [58].
- Functional Analysis:
- Heterologous Expression: Express the gene in a system like E. coli or yeast to study its substrate specificity (e.g., for malate, glutamate, 2-oxoglutarate) using uptake assays with radiolabeled or stable isotopes [59].
- Metabolite Analysis: Compare the levels of photorespiratory and TCA cycle intermediates (malate, 2-oxoglutarate, glutamate, glycine, serine) in the mutant versus wild-type under stress conditions using GC-MS or LC-MS [58].
Research Reagent Solutions
Table: Essential research reagents and their applications in CCM and photorespiration studies.
| Reagent / Tool | Function / Application | Key Characteristics / Target |
|---|---|---|
| Chlamydomonas CLiP Mutant Library [58] | Provides ready-to-use insertion mutants for reverse genetics. | Targets include LCI20, CIA5 (CCM master regulator), HLA3 (bicarbonate transporter). |
| LCI20 Transporter | Chloroplast envelope glutamate/malate exchanger; links photorespiration with CCM energy supply [58] [59]. | Essential for growth during acute shift to very low COâ‚‚; a key node for coordination. |
| CIA5 Transcription Factor | Master regulator controlling the induction of both CCM and photorespiratory genes in response to low COâ‚‚ [58]. | cia5 mutants are unable to induce CCM and grow poorly in low COâ‚‚ air. |
| HLA3 Transporter | ATP-binding cassette (ABC) transporter at the plasma membrane; actively imports bicarbonate under very low COâ‚‚ conditions [58]. | Requires ATP, highlighting the energy demand of CCMs. |
| Carbonic Anhydrases (e.g., CAH3) | Enzymes that interconvert COâ‚‚ and bicarbonate, facilitating COâ‚‚ diffusion and concentration near Rubisco [62] [58]. | CAH3 is located in the thylakoid lumen and is crucial for dehydrating bicarbonate to COâ‚‚ in the pyrenoid. |
| Fluorescent Protein Tags (mVenus) | Used for protein localization studies (e.g., to confirm chloroplast envelope localization) [58]. | Fused to the protein of interest (e.g., LCI20-mVenus) and expressed under a constitutive promoter. |
| 2% Oxygen Atmosphere | Experimental condition to suppress Rubisco oxygenase activity and photorespiration [58]. | Serves as a control to determine if a mutant's phenotype is specifically linked to photorespiration. |
Table: Quantitative data on growth and glycolate excretion in Chlamydomonas mutants under different COâ‚‚ conditions (based on [58]).
| Strain / Genotype | Growth in High COâ‚‚ | Growth in Low COâ‚‚ | Growth in Very Low COâ‚‚ | Glycolate Excretion at Low COâ‚‚ |
|---|---|---|---|---|
| Wild Type | Normal | Normal | Normal | Low to Moderate |
lci20 mutant |
Normal | Slightly Impaired | Severely Impaired | Increased |
cia5 mutant |
Normal | No Growth | No Growth | High (due to lack of CCM) |
| Photorespiratory mutant (e.g., GlcDH) | Normal | Impaired (in air) | Impaired (in air) | Very High |
Table: Key genetic modifications for optimizing photosynthesis and reducing photorespiration (based on [62]).
| Genetic Target | Type of Modification | Expected Physiological Outcome |
|---|---|---|
| C4 Enzymes (PEPC, NADP-ME) | Introduce from C4 plants into C3 organisms. | Creates localized COâ‚‚ concentration around Rubisco, suppressing oxygenase activity [62]. |
| Rubisco Specificity | Engineer Rubisco large and small subunits. | Increase carboxylation efficiency over oxygenation (Sc/o value) [62] [60]. |
| Cyanobacterial COâ‚‚ pumps | Introduce Bicarbonate Transporters (BicA, SbtA). | Enhance active uptake of inorganic carbon into the cell [62]. |
| Chloroplast Electron Transport | Overexpress Cytochrome b6f complex or FNR. | Increase ATP and NADPH production to meet higher energy demands of CCMs [62]. |
| Synthetic Photorespiratory Bypass | Introduce alternative pathways (e.g., glycolate catabolic pathways from bacteria). | Reduce carbon and energy loss by more efficiently recycling 2-PG [62] [60]. |
Validation and Comparative Analysis of CCM Performance Across Algal Systems
FAQ: Algal Carbon Concentrating Mechanisms (CCMs)
Q1: What is the fundamental purpose of a CCM in photosynthetic algae? Aquatic environments present a major challenge for photosynthesis because COâ‚‚ diffuses 10,000 times more slowly in water than in air, and the primary carboxylating enzyme, RuBisCO, has a low affinity for COâ‚‚ and is prone to oxygenase activity leading to photorespiration [63]. The COâ‚‚ concentrating mechanism (CCM) is a biological adaptation that actively increases the intracellular concentration of COâ‚‚ at the site of RuBisCO. This enhances photosynthetic efficiency by promoting carboxylation and suppressing photorespiration [64] [4].
Q2: What are the main types of CCMs found in algae? Algae primarily utilize two types of CCMs:
- Biophysical CCMs: Rely on the active transport of inorganic carbon (Ci; COâ‚‚ and HCO₃â») across cellular membranes and the interconversion of Ci species by carbonic anhydrase (CA) to elevate COâ‚‚ levels around RuBisCO [2] [64].
- Biochemical CCMs: Often compared to C4 photosynthesis in plants, these mechanisms involve the biochemical fixation of Ci into C4 organic acids, which are then transported and decarboxylated to release COâ‚‚ near RuBisCO [2].
Q3: How do model algal systems differ in their CCM strategies? The coordination and dominance of biophysical versus biochemical CCM components vary significantly between major model algae, as summarized in the table below.
Table 1: Comparison of CCM Strategies in Model Algae
| Model Organism | Dominant CCM Strategy | Key Components and Features | Environmental Plasticity |
|---|---|---|---|
| Chlamydomonas reinhardtii | Primarily Biophysical [7] [9] | Well-characterized Ci transporters (HLA3, LCIA), pyrenoid, multiple carbonic anhydrases (CAH1, CAH3, CAH6) [9] [63]. | CCM and photorespiration genes co-induced by low COâ‚‚ via master regulator CIA5 [7]. |
| Ulva prolifera | Mixed; Biophysical dominates, Biochemical supports [2] [3] | Biophysical can compensate for ~100% of carbon fixation; biochemical CCM contributes ~50% when biophysical is inhibited. Involves C4 enzymes (PEPCK) [2] [4]. | High degree of coordination; the two CCM types complement and reinforce each other under stress [2]. |
| Diatoms (e.g., Thalassiosira weissflogii) | Mixed; can utilize both [2] | Biochemical CCM may dominate under specific stresses (e.g., Zn-limitation), involving PEPC and PEPCK enzymes [2]. | Exhibits plasticity, shifting CCM contribution in response to environmental changes like Zn availability [2]. |
Q4: What is the relationship between the CCM and photorespiration? While it was long thought that an operational CCM would suppress photorespiration by saturating RuBisCO with COâ‚‚, recent evidence in Chlamydomonas shows that photorespiration remains active even when the CCM is induced under low COâ‚‚ conditions [7]. The two processes are co-regulated, and photorespiration may play a crucial role in managing metabolic flux during acclimation.
Troubleshooting Common Experimental Challenges
Challenge 1: Differentiating between Biophysical and Biochemical CCM Contributions Problem: It is experimentally difficult to distinguish the individual contributions of biophysical and biochemical CCMs to total carbon fixation in an algal cell [2]. Solution: Use specific enzyme inhibitors in combination with photosynthetic measurements.
- Inhibitor for Biophysical CCM: Ethoxyzolamide (EZ), an inhibitor of carbonic anhydrase (CA). A typical working concentration is 50 µM [2].
- Inhibitor for Biochemical CCM: 3-Mercaptopicolinic acid (MPA), an inhibitor of phosphoenolpyruvate carboxykinase (PEPCK). A typical working concentration is 1.5 mM [2].
- Protocol: Measure the rate of photosynthetic Oâ‚‚ evolution or carbon fixation in control cells and compare it to the rates after adding EZ, MPA, or both. The percentage reduction in photosynthesis indicates the relative contribution of each pathway. For example, in Ulva, EZ inhibition of the biophysical CCM led to a ~50% drop in fixation, which was compensated by increased activity of the biochemical CCM [2].
Challenge 2: Assessing CCM Induction and Functionality Problem: Determining if an algal strain has an inducible or constitutively active CCM. Solution: Perform a photosynthetic inorganic carbon (P-C) response curve [3] [4].
- Protocol:
- Acclimate algal cultures to both high COâ‚‚ (e.g., 2-5%) and low/ambient COâ‚‚ (0.04%) conditions.
- Deplete the cells of endogenous Ci by transferring them to a Ci-free buffer.
- Using an O₂ electrode, measure the photosynthetic rate while sequentially adding known concentrations of NaHCO₃ to the medium.
- Plot the photosynthetic rate against the DIC or COâ‚‚ concentration. A lower half-saturation constant (Kâ‚/â‚‚) in low-COâ‚‚-acclimated cells indicates a higher affinity for Ci and the induction of an active CCM [4].
Challenge 3: Unexpected Localization of CCM Components Problem: When expressing algal CCM genes in heterologous systems (e.g., tobacco or Arabidopsis), proteins may not localize to the correct compartment [9]. Solution: Always verify subcellular localization experimentally.
- Protocol: Fuse the gene of interest to a fluorescent reporter (e.g., GFP, mVenus) and express it under a constitutive promoter. Confirm localization using confocal microscopy. For instance, the putative Ci transporter LCI1 from Chlamydomonas was found on the plasma membrane, but for chloroplast function in a higher plant, it required the addition of a chloroplast transit peptide for retargeting [9].
The Scientist's Toolkit: Key Research Reagents
Table 2: Essential Reagents for Studying Algal CCMs
| Reagent / Material | Function / Application | Key Details / Example |
|---|---|---|
| Ethoxyzolamide (EZ) | Inhibitor of carbonic anhydrase (CA); used to suppress the biophysical CCM. | Inhibits both extracellular and intracellular CA activity [2]. |
| 3-Mercaptopicolinic Acid (MPA) | Inhibitor of PEP carboxykinase (PEPCK); used to suppress the biochemical (C4-like) CCM. | Effective for probing C4 metabolism in diatoms and Ulva [2]. |
| Acetazolamide (AZ) | Specific inhibitor of external, periplasmic carbonic anhydrase. | Used to dissect the role of external CA versus internal CA [2]. |
| Clark-type Oâ‚‚ Electrode | Measuring photosynthetic oxygen evolution rates. | Essential for generating P-C curves and assessing CCM activity [2] [3]. |
| HLA3, LCIA, LCI1 Genes | Molecular components (Ci transporters) for engineering CCMs. | HLA3 and LCIA are HCO₃⻠transporters confirmed to function in Xenopus oocytes [9]. |
| EPYC1 Protein | Essential linker protein for pyrenoid formation. | A low-complexity protein that packages RuBisCO into the pyrenoid microcompartment [63]. |
Experimental Workflow & Signaling Pathways
The following diagram illustrates a generalized experimental workflow for dissecting CCM contributions in algae, based on methodologies applied to Ulva prolifera [2].
The diagram below summarizes the current understanding of the coordination between biophysical and biochemical CCMs and their relationship with photorespiration, as revealed in Chlamydomonas and Ulva [2] [7].
Technical Support Center
Troubleshooting Guides
Guide 1: Troubleshooting Experimental Challenges in CCM Mutant Phenotyping
1. Identify the Problem: Impaired growth in algal strains during transition from high CO2 (H-CO2) to very low CO2 (VL-CO2) conditions, specifically in mutants with defects in carbon-concentrating mechanism (CCM) components [7].
2. List All Possible Explanations:
- Defective CCM induction
- Disrupted photorespiratory pathway
- Incorrect growth conditions (CO2 level, light intensity)
- Issues with mitochondrial energy trafficking
- Problematic culture media
3. Collect the Data:
- Verify CO2 levels in growth chambers using a CO2 analyzer.
- Check light intensity with a photometer.
- Confirm genotype of mutant strains via PCR and sequencing.
- Analyze expression levels of key CCM genes (e.g.,
HLA3,LCIA,CCP1/2,CAH3) via RT-qPCR in mutant vs. wild-type under inducing conditions [9] [7]. - Monitor cell growth by measuring optical density and cell count.
4. Eliminate Some Possible Explanations:
- If CO2 levels and light intensity are confirmed correct, eliminate "incorrect growth conditions."
- If genotyping confirms the mutation, focus on downstream functional consequences.
5. Check with Experimentation:
- Photorespiration Test: Compare mutant growth under VL-CO2 at 21% O2 (photorespiratory) versus 2% O2 (suppresses photorespiration). Growth rescue at low O2 suggests a photorespiratory defect [7].
- Glycolate Excretion Assay: Measure glycolate in the culture medium. Elevated excretion indicates a blockage in photorespiratory metabolism [7].
- Ci Uptake Assay: Perform Hâ´CO₃⻠uptake assays to directly test for defects in inorganic carbon transport [9].
6. Identify the Cause: Based on experimental results. For example, if a mutant shows growth rescue at low O2 and high glycolate excretion, the cause is likely a defective photorespiratory pathway, not a primary CCM defect [7].
Guide 2: Troubleshooting Localization of Algal CCM Components in Heterologous Systems
1. Identify the Problem: Heterologously expressed algal CCM protein (e.g., CAH3, CAH6, LCI1) fails to localize to the correct organelle in a model plant like tobacco or Arabidopsis [9].
2. List All Possible Explanations:
- Missing or inefficient native algal transit peptide
- Incorrect subcellular targeting information in algal protein sequence
- Protein instability or degradation in the host system
- Incompatibility with the host's organellar import machinery
3. Collect the Data:
- Use confocal microscopy to visualize the actual location of the fluorescently tagged protein.
- Perform immunoblotting on fractionated cellular components to confirm protein presence and size.
- Use bioinformatic tools to predict the native algal protein's targeting signals.
4. Eliminate Some Possible Explanations:
- If the protein is detected at full-length but in the wrong location, eliminate "protein degradation."
- If bioinformatics strongly predict a chloroplast transit peptide, the issue is likely not "incorrect targeting information."
5. Check with Experimentation:
- Transit Peptide Swapping: Fuse the algal protein to a well-characterized, strong transit peptide from the host plant (e.g., from Arabidopsis
RBCSfor chloroplast targeting). Re-check localization [9]. - Sequence Analysis: Compare the algal transit peptide to known functional transit peptides from the host to identify key motif deficiencies.
6. Identify the Cause: If re-targeting with a host-specific transit peptide successfully localizes the protein to the correct organelle, the cause is the inefficiency of the native algal transit peptide in the heterologous system [9].
Frequently Asked Questions (FAQs)
Q1: We are trying to improve photosynthetic efficiency in a C3 plant by introducing algal CCM components. Why does expressing a single Ci transporter, like HLA3 or LCIA, not lead to enhanced growth?
A1: The algal CCM is a complex, multi-component system. Introducing a single transporter is insufficient because it requires coordination with other elements, including specific carbonic anhydrases (e.g., CAH3 in the pyrenoid) and a structured pyrenoid microcompartment around RuBisCO. Stacking multiple components—transporters, CAs, and pyrenoid proteins—is likely necessary to create a functional, synergistic system that significantly increases CO2 concentration at the RuBisCO site [9].
Q2: What is the relationship between the CCM and photorespiration in Chlamydomonas? Does an active CCM completely suppress photorespiration? A2: No, recent evidence indicates that the CCM and photorespiration operate jointly under low CO2 conditions. The CCM reduces but does not eliminate photorespiration. Photorespiration remains active, serving as a detoxification pathway for 2-phosphoglycolate (2-PG). Furthermore, glycolate excretion acts as a safety valve to prevent the toxic accumulation of photorespiratory intermediates when the CCM is operational [7].
Q3: Our lab identified a putative Ci transporter in a new algal species. What is a robust experimental method to confirm its function? A3: A reliable method is heterologous expression and functional characterization in Xenopus laevis oocytes. By injecting the transporter's mRNA into oocytes and conducting Hâ´CO₃⻠uptake assays, you can directly measure and quantify the protein's ability to transport inorganic carbon. This approach provides direct evidence of transport function independent of the native algal background [9].
Q4: We are observing high variability in our measurements of synaptic coupling strength from cross-correlation histograms (CCHs) in neuronal data related to E:I balance. How can we improve the reliability of our quantifications? A4: Ensure you have a sufficiently large dataset. CCH analysis requires substantial spiking data for statistical power. For dynamic measurements, consider aggregating data from time windows with similar behavioral or physiological states (e.g., grouping by percentiles of LFP aperiodic slope) to build up enough spikes for stable CCH calculation. The scale of the recording is critical; high-density silicon probes that capture many single units simultaneously are advantageous [65].
Experimental Protocols & Data Presentation
Table 1: Key CCM Components inChlamydomonas reinhardtii: Functions, Localizations, and Mutant Phenotypes
| Gene | Putative Function | Native Localization (Chlamydomonas) | Localization in Tobacco (Heterologous) | Mutant Phenotype (Under Low CO2) |
|---|---|---|---|---|
| HLA3 | Ci (HCO₃â») uptake into cytosol [9] | Plasma Membrane [9] | Not specified | High-COâ‚‚-Requiring (HCR); reduced Ci accumulation [9] |
| LCI1 | Ci uptake into cytosol [9] | Plasma Membrane [9] | Chloroplast (when fused with host transit peptide) [9] | Promotes HCO₃⻠uptake when overexpressed [9] |
| LCIA | Ci transport to chloroplast stroma [9] | Chloroplast Envelope [9] | Same as native (Chloroplast Envelope) [9] | HCR; reduced Ci accumulation [9] |
| CCP1/2 | Putative Ci transport [9] | Mitochondria [9] | Same as native (Mitochondria) [9] | Role in Ci transport supported by RNAi [9] |
| CAH3 | Dehydration of HCO₃⻠to CO₂ near RuBisCO [9] | Thylakoid Lumen [9] | Incorrect (required retargeting) [9] | Overaccumulation of Ci [9] |
| CAH6 | Recapture of leaking COâ‚‚ [9] | Chloroplast Stroma [9] | Incorrect (required retargeting) [9] | No published mutant [9] |
| LCIB/C | COâ‚‚ uptake/trapping; pyrenoid periphery [9] | Pyrenoid Periphery [9] | Same as native (Pyrenoid Periphery) [9] | Lethal under air-level COâ‚‚ (LCIB) [9] |
Table 2: Dynamic Synaptic Coupling and LFP Spectral Characteristics in Rat mPFC
| Behavioral State in Restaurant Row Task | Excitatory Synaptic Strength (from CCHs) | Inhibitory Synaptic Strength (from CCHs) | E:I Ratio | LFP Aperiodic Slope (1/f Exponent) | Broadband Spectral Power |
|---|---|---|---|---|---|
| Wait Zone (Waiting during delay) | Stronger [65] | Stronger [65] | Dynamic | Higher (steeper) [65] | Measured [65] |
| Reward Zone (Consuming reward) | Stronger [65] | Stronger [65] | Dynamic | Lower (shallower) [65] | Measured [65] |
| Correlation with E:I Balance | --- | --- | Strong inverse relationship with Broadband Power [65] | Mild positive correlation (opposite to hypothesis) [65] | Strongly correlated with E:I balance [65] |
Protocol 1: Functional Characterization of a Putative Bicarbonate Transporter inXenopusOocytes
Methodology:
- Vector Construction: Clone the full-length coding sequence (CDS) of the putative transporter (e.g.,
HLA3,LCIA) into a high-expression vector suitable for in vitro transcription. - mRNA Synthesis: Perform in vitro transcription to generate capped mRNA from the linearized plasmid.
- Oocyte Injection: Manually inject a defined amount (e.g., 50 ng) of mRNA into stage V-VI Xenopus laevis oocytes. Maintain control oocytes injected with nuclease-free water.
- Incubation: Incubate oocytes for 2-4 days at a defined temperature to allow for protein expression.
- Hâ´CO₃⻠Uptake Assay:
- Individually place oocytes in a solution containing radioactive Hâ´CO₃â».
- Incubate for a precise time (e.g., 30 minutes).
- Rapidly wash oocytes with ice-cold, non-radioactive buffer to terminate uptake.
- Lysate individual oocytes and quantify radioactivity using a liquid scintillation counter.
- Data Analysis: Compare Hâ´CO₃⻠uptake in mRNA-injected oocytes versus water-injected controls. Statistically significant higher uptake in mRNA-injected oocytes confirms transport function [9].
Protocol 2: Differentiating CCM and Photorespiration Defects in Algal Mutants
Methodology:
- Pre-culture: Grow wild-type and mutant algal strains under high CO2 (e.g., 2-5%) to mid-log phase.
- Experimental Setup: Wash and resuspend cells in fresh, low-carbon media. Divide cultures into four conditions:
- Condition A: VL-CO2 (0.01%) + 21% O2 (Photorespiratory)
- Condition B: VL-CO2 (0.01%) + 2% O2 (Non-Photorespiratory)
- Condition C: H-CO2 (2%) + 21% O2 (Control)
- Condition D: H-CO2 (2%) + 2% O2 (Control)
- Growth Monitoring: Track culture growth for 3-7 days by measuring optical density at 750 nm (OD₇₅₀) and cell counts using a hemocytometer or automated cell counter.
- Glycolate Measurement: Collect culture supernatant from different time points. Quantify excreted glycolate using a colorimetric enzyme assay kit or via HPLC.
- Interpretation:
- CCM Defect (e.g.,
cia5): No growth in both VL-CO2 conditions (A & B), regardless of O2 level [7]. - Photorespiratory Defect (e.g.,
lci20): Impaired growth in VL-CO2 at 21% O2 (A), but growth is rescued or significantly improved in VL-CO2 at 2% O2 (B). This is often accompanied by elevated glycolate excretion [7].
- CCM Defect (e.g.,
Diagrams of Signaling Pathways and Workflows
CCM-Photorespiration Coordination
E:I Balance Experimental Workflow
Mutant Phenotyping Logic
The Scientist's Toolkit: Research Reagent Solutions
| Item | Function/Application | Example/Note |
|---|---|---|
| Chlamydomonas CLiP Mutant Library | Source of targeted knockout mutants for genes of interest (e.g., lci20) [7]. |
Essential for reverse genetics studies [7]. |
| CIA5 Antibody | Verifies the presence/absence of the master regulator protein in mutant strains [7]. | Critical for confirming cia5 phenotype at protein level [7]. |
| LCI20 Antibody | Confirms knockout/complementation and studies protein localization [7]. | Used to validate lci20 mutant and complemented lines [7]. |
| Heterologous Expression System (Xenopus oocytes) | Functional validation of putative Ci transporters (e.g., HLA3, LCIA) via Hâ´CO₃⻠uptake assays [9]. |
Provides direct evidence of transport capability [9]. |
| Specific Cis-Elements/Chimeric Reporters | Identifies and characterizes gene promoters responsive to CO2 levels (CCM regulation). | Used to study transcriptional regulation of CCM genes. |
| High-Density Silicon Neural Probes | Enables large-scale simultaneous recording of LFP and single units for E:I balance studies [65]. | Key for obtaining sufficient data for CCH analysis [65]. |
| Colorimetric Glycolate Assay Kit | Quantifies glycolate excretion in culture media, a key indicator of photorespiratory status [7]. | Standardizes measurement of photorespiratory flux [7]. |
Frequently Asked Questions (FAQs)
FAQ 1: Why does my strain show high biomass accumulation but unexpectedly low lipid productivity, despite an induced CCM? A fully induced CCM does not automatically guarantee enhanced lipid biosynthesis. The carbon fixed by the CCM must be actively channeled into the lipid synthesis pathway. Research shows that key enzymes for fatty acid biosynthesis, such as acetyl-CoA carboxylase (ACC) subunits, localize near the pyrenoid under COâ‚‚ limitation. If the pyrenoid is functionally disrupted, it can impair the transfer of fixed carbon to lipids, leading to the observed discrepancy. Ensure your cultivation strategy does not create a pyrenoid dysfunction and check for genetic mutations affecting pyrenoid structure [8].
FAQ 2: We confirmed the expression of CCM genes in our strain, but the overall biomass productivity has dropped. What is the energy cost of running a CCM? The operation of a CCM is energetically expensive. Energy is required for active inorganic carbon (Ci) uptake and for maintaining ion gradients. This energy (ATP and reducing power) is diverted from biomass synthesis. The trade-off between carbon concentration and energy expenditure is a critical factor. Modeling studies suggest that parameters like bicarbonate pumping cost and kinetics significantly influence whether the net effect of a CCM is positive for growth. You may be observing this trade-off in your strain. Optimizing light intensity to ensure sufficient energy production can help mitigate this issue [66] [56] [7].
FAQ 3: During the acclimation from high to low COâ‚‚, our culture exhibits poor growth and excretes glycolate. Is this normal? Yes, this can be a normal transitional phase. During acclimation to low COâ‚‚, the CCM is not yet fully operational, leading to elevated Rubisco oxygenase activity and photorespiration. Glycolate is a key intermediate of photorespiration, and its excretion is a mechanism to avoid the toxic buildup of photorespiratory metabolites before the CCM is fully induced and suppresses photorespiration. This process is active in strains like Chlamydomonas reinhardtii. Monitoring glycolate levels can serve as a useful indicator of the acclimation status [7].
FAQ 4: Can a CCM function effectively in a non-canonical alga that lacks structures like a pyrenoid? Yes, evidence supports the existence of functional, non-canonical CCMs. Studies on the red alga Cyanidioschyzon merolae, which lacks a pyrenoid and the ability to take up external bicarbonate, demonstrate that a minimal CCM can operate. Mathematical modeling indicates that features such as cytosolic pH, cell radius, and COâ‚‚ membrane permeability are essential. If you are working with a non-model production strain, it may possess such a minimal CCM, and characterizing these fundamental parameters is key to understanding and optimizing its performance [66].
Troubleshooting Guides
Table 1: Troubleshooting Low Biomass Output Under Induced CCM Conditions
| Symptom | Possible Cause | Recommended Experimentation & Validation |
|---|---|---|
| Low biomass and low lipid content | Energy drain from CCM operation outweighs carbon gain. | 1. Measure photosynthetic parameters: Quantify Oâ‚‚ evolution or PSII efficiency under low COâ‚‚. 2. Optimize light intensity: Increase light to meet the additional ATP demand of the CCM [7]. |
| High biomass but low lipid yield | Disconnect between CCM and lipid biosynthesis pathway; dysfunctional pyrenoid. | 1. Visualize the pyrenoid: Use microscopy to confirm pyrenoid integrity. 2. Localize ACCase: Verify that acetyl-CoA carboxylase subunits are present at the pyrenoid periphery under low COâ‚‚ [8]. |
| Slow growth after COâ‚‚ shift | Inefficient CCM induction or impaired photorespiratory metabolism. | 1. Monitor gene expression: Track expression of key CCM genes (e.g., HLA3, LCIA) and photorespiratory genes (e.g., LCI20) via qPCR during transition [7]. 2. Assay glycolate excretion: Measure extracellular glycolate as a marker of photorespiratory activity [7]. |
| Poor Ci uptake in a non-model strain | Non-canonical CCM with unknown transporters; suboptimal cultivation conditions. | 1. Characterize affinity: Measure the whole-cell affinity for CO₂ (Kₘ) and compare it to the affinity of its purified Rubisco. 2. Model the system: Use compartmental modeling to identify essential features like cytosolic pH and membrane permeability that could be optimized [66]. |
Table 2: Quantifiable Impact of CCM Modulation on Biomass and Lipids in Various Algal Strains
| Algal Strain | Experimental Modification / Condition | Impact on Biomass | Impact on Lipids / Triacylglycerol (TAG) | Key Finding |
|---|---|---|---|---|
| Chlamydomonas reinhardtii | Pyrenoid-disrupted mutant | Not specified | Impaired fatty acid and TAG biosynthesis [8] | Pyrenoid functionality is essential for channeling fixed carbon into lipids. |
| Chlorella vulgaris / C. sorokiniana | Overexpression of bacterial carbonic anhydrase (MlCA) | Enhanced biomass production with 1% COâ‚‚ supply [67] | Increased lipid accumulation [67] | Enhanced carbon capture via CA directly boosts biomass and lipid yields. |
| Chlamydomonas reinhardtii lci20 mutant (defective in photorespiration) | Transition from high to very-low COâ‚‚ | Severely impaired growth during transition [7] | Not specified | Photorespiration is essential for managing metabolic flux during CCM induction. |
| Neochloris oleoabundans | Mixotrophic cultivation (vs. autotrophy) | Elevated biomass production [68] | Overproduction of lipids [68] | Mixed carbon sources enhance overall carbon accrual and storage compound synthesis. |
Experimental Protocols for Key Validations
Protocol 1: Validating CCM and Lipid Pathway Coordination via ACCase Localization
Objective: To confirm that the carbon concentrated by the CCM is directly utilized for lipid biosynthesis by visualizing the co-localization of ACCase with the pyrenoid.
Materials:
- Strains: Your production strain of interest, cultured under high COâ‚‚ (e.g., 2-5%) and after acclimation to low COâ‚‚ (air level, 0.04%).
- Reagents: Fixative (e.g., formaldehyde), permeabilization buffer (e.g., Triton X-100), primary antibody against ACCase, fluorescently-labeled secondary antibody, mounting medium with DAPI or similar for nuclear/chloroplast staining.
- Equipment: High-resolution fluorescence or confocal microscope.
Methodology:
- Culture & Induction: Grow two batches of algae. Maintain one under high COâ‚‚ and shift the other to low COâ‚‚ conditions for at least 24 hours to fully induce the CCM.
- Fixation and Permeabilization: Harvest cells by gentle centrifugation. Resuspend in fixative for 15-30 minutes. Pellet cells and wash with buffer. Permeabilize cells with a mild detergent solution.
- Immunostaining: Incubate cells with the primary antibody against ACCase. Wash thoroughly to remove unbound antibody. Incubate with the fluorescent secondary antibody. Include a control sample that omits the primary antibody to check for non-specific binding.
- Microscopy and Analysis: Mount the cells and visualize under a microscope. Under low COâ‚‚ conditions, ACCase should form punctate condensates at the periphery of the pyrenoid (visualized as a dense region within the chloroplast). Under high COâ‚‚, this localization should be diffuse throughout the chloroplast stroma [8].
Protocol 2: Quantifying the Functional Outcome of CCM via Glycolate Excretion Profiling
Objective: To assess the operational status of the CCM and its suppression of photorespiration by measuring glycolate excretion during acclimation to low COâ‚‚.
Materials:
- Strains: Your production strain.
- Reagents: Colorimetric glycolate assay kit (e.g., involving periodate oxidation), cell culture medium filtrate.
- Equipment: Centrifuge, microplate reader or spectrophotometer.
Methodology:
- Acclimation Time-Course: Shift a high-COâ‚‚-grown culture to low COâ‚‚ conditions. Collect culture samples at specific time points (e.g., 0, 2, 6, 12, 24, 48 hours).
- Sample Preparation: At each time point, centrifuge the culture sample to pellet cells. Carefully filter the supernatant (0.2 µm) to remove all cells and debris.
- Glycolate Assay: Follow the manufacturer's instructions for the glycolate assay kit. Typically, this involves reacting the cell-free supernatant with specific reagents to produce a colored product whose absorbance is proportional to the glycolate concentration.
- Data Interpretation: A successful CCM induction is characterized by a transient peak in glycolate excretion, which subsequently declines as the CCM becomes fully operational and suppresses Rubisco oxygenase activity. A persistently high level of glycolate suggests an incomplete or dysfunctional CCM [7].
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Key Reagents for Investigating CCM and Biomass Coordination
| Reagent / Material | Function in CCM-Biomass Research | Example from Literature |
|---|---|---|
| Antibody against Acetyl-CoA Carboxylase (ACCase) | To visualize the spatial coordination between the CCM and lipid biosynthesis pathways via immunofluorescence. | Used to demonstrate re-localization of ACCase subunits to the pyrenoid periphery under COâ‚‚ limitation in Chlamydomonas [8]. |
| Glycolate Assay Kit | To quantify photorespiratory flux and indirectly monitor the efficiency of CCM in suppressing Rubisco oxygenase activity. | Employed to profile glycolate excretion during the acclimation of Chlamydomonas to low COâ‚‚ conditions [7]. |
| Expression Vector with High-Activity CA Gene (e.g., MlCA) | To genetically enhance the carbon fixation capacity of the CCM and test its impact on downstream biomass and lipid production. | The Mesorhizobium loti CA gene (MlCA) was transformed into Chlorella spp., leading to enhanced COâ‚‚ fixation and lipid accumulation [67]. |
| Cultivation System for Mixotrophy | To provide both organic and inorganic carbon, boosting overall biomass and altering metabolic fluxes towards products like lipids and exo-polysaccharides. | Used with Neochloris oleoabundans to achieve elevated biomass and lipid production compared to autotrophic conditions [68]. |
Signaling Pathways and Experimental Workflows
CCM Induction and Metabolic Integration Pathway
Experimental Workflow for CCM-Biomass Validation
FAQ: Troubleshooting Common Experimental Challenges
FAQ 1: My heterologous CCM genes have been successfully integrated into the algal host, but I'm not observing the expected enhancement in carbon fixation. What could be wrong?
This is a common issue often stemming from improper coordination between the transferred biophysical CCM and the host's native biochemical CCM (Calvin Cycle). First, verify that the transferred genes are being expressed at the protein level via Western blot. Second, assess the physiological context; the host's biochemical CCM might be downregulating in response to the new biophysical CCM. Inhibiting the biophysical CCM with ethoxyzolamide can trigger a compensatory increase in biochemical CCM activity, and vice-versa. This complementary relationship must be balanced for optimal function [35].
FAQ 2: The algal strain shows poor growth and viability after the introduction of a cross-species iModulon. How can I resolve this?
Suboptimal performance after cross-species transfer is frequently due to metabolic burden or improper interaction with the host's native systems. Adaptive Laboratory Evolution (ALE) is a highly effective strategy to overcome this. By subjecting the engineered strain to prolonged growth under selective pressure (e.g., low COâ‚‚ conditions), you can force the host genome to accumulate compensatory mutations that optimize the new function. This approach has been successfully used to optimize cellular functions transferred from Pseudomonas species to E. coli [69].
FAQ 3: I am encountering inconsistent results with my CRISPR-Cas editing system in microalgae when introducing heterologous CCM components. What are the key factors to check?
The efficiency of CRISPR-driven engineering in microalgae is highly dependent on several species-specific factors:
- Cas Protein and PAM Requirements: The canonical SpCas9 (requiring an NGG PAM) may not be optimal for all algal species. Consider testing smaller orthologs like Cas12a (which recognizes T-rich PAMs like TTTV) or high-fidelity variants to improve efficiency and reduce off-target effects [70].
- Delivery Method: The rigid cell walls of many microalgae are a major barrier. If electroporation or biolistics are yielding low efficiency, optimize the protocol or explore novel delivery vehicles.
- Cellular Repair Machinery: Microalgae often have inefficient Homology-Directed Repair (HDR). Using CRISPR-based editors that do not rely on HDR, such as base editors (CBEs, ABEs) or prime editors (PEs), can lead to more precise and successful edits [70].
â–¼ Key Reagent Solutions for CCM Transfer Experiments
Table 1: Essential research reagents and their applications in cross-species CCM transfer.
| Reagent / Tool | Primary Function | Application in CCM Experiments |
|---|---|---|
| iModulon Analysis [69] | Identifies complete set of independently modulated genes for a cellular function. | Precisely defines all genetic components (including unannotated genes) required for transferring a biophysical CCM from a donor species. |
| CRISPR-dCas9 (CRISPRa/i) [70] | Enables tunable gene activation (a) or interference (i) without DNA cleavage. | Fine-tunes the expression of transferred CCM genes or host native biochemical CCM genes (e.g., RuBisCO) to optimize coordination. |
| Base Editors (CBEs, ABEs) [70] | Facilitates single-nucleotide conversions without double-strand breaks. | Creates precise point mutations in host genome to improve compatibility with the transferred CCM, minimizing metabolic distress. |
| Ethoxyzolamide [35] | Inhibitor of carbonic anhydrase, a key enzyme in biophysical CCMs. | Used experimentally to inhibit the transferred biophysical CCM, allowing researchers to probe the compensatory response of the biochemical CCM. |
| Adaptive Laboratory Evolution (ALE) [69] | Optimizes cellular functions through long-term cultivation under selective pressure. | Applied post-transfer to force the host algae to adapt and optimize the newly acquired CCM function for enhanced growth and carbon fixation. |
â–¼ Quantitative Analysis of CCM Coordination in Engineered Algae
Table 2: Experimentally measured compensation between biophysical and biochemical CCMs in Ulva prolifera after inhibition. Data adapted from Zhang et al. (2025b) [35].
| Experimental Condition | CCM Type Targeted | Observed Reduction in Carbon Fixation | Compensatory Increase from Alternate CCM |
|---|---|---|---|
| Biophysical CCM Inhibited (Ethoxyzolamide) | Biophysical | Reduced | ~50% compensation via Biochemical CCM |
| Biochemical CCM Inhibited (3-mercaptopicolinic acid) | Biochemical | Reduced | ~100% compensation via Biophysical CCM |
â–¼ Detailed Experimental Protocol: iModulon-Based Transfer and Optimization
This protocol outlines the methodology for cross-species transfer of a Carbon Concentration Mechanism (CCM) using an iModulon-based approach, followed by host optimization via Adaptive Laboratory Evolution (ALE).
Step 1: Identification of the CCM iModulon
- Method: Apply Independent Component Analysis (ICA) to large transcriptomic datasets from the donor organism (e.g., a cyanobacterium with a desired CCM) available in databases like iModulonDB.
- Objective: Identify the iModulon whose activity correlates strongly with low-COâ‚‚ conditions. This iModulon represents the set of co-regulated genes constituting the functional CCM unit, including core and accessory genes [69].
Step 2: Refactoring and Cloning
- Method: Synthesize the genes identified in the CCM iModulon. Where possible, preserve their native genetic arrangement (e.g., operon structure) to ensure balanced expression.
- Cloning: Assemble these genes into a suitable expression vector containing a promoter (e.g., an inducible Trc promoter) that functions in the algal host [69].
Step 3: Transformation and Screening
- Delivery: Introduce the constructed plasmid into the host microalgae using an optimized transformation method (e.g., electroporation, biolistics).
- Screening: Select successful transformants using the plasmid's antibiotic resistance marker. Confirm the presence and expression of the transferred genes via PCR and RT-qPCR [69].
Step 4: Functional Validation and ALE
- Validation: Grow validated strains in sealed photobioreactors under low-COâ‚‚ conditions. Measure carbon fixation rates and intracellular pH shifts compared to a wild-type control.
- ALE: Inoculate the validated strain into a minimal medium with low COâ‚‚ availability as the selective pressure. Serial passage the culture for hundreds of generations. Monitor for the evolution of faster-growing mutants [69].
- Post-ALE Analysis: Sequence the genomes of evolved, high-performing strains to identify mutations that contributed to the optimized phenotype [69].
â–· Visualizing the Experimental Workflow and CCM Coordination
Experimental Workflow for CCM Transfer
CCM Coordination Mechanism
In algal research, the CO2-concentrating mechanism (CCM) is a critical adaptive strategy that enables efficient photosynthesis despite the kinetic limitations of the key enzyme Rubisco [56]. This mechanism exists in two principal forms: the biophysical CCM, which relies on active transport of inorganic carbon (Ci) and the catalytic activity of carbonic anhydrases, and the biochemical CCM (or C4-like metabolism), which involves the synthesis and decarboxylation of C4 acid intermediates [2]. Optimal coordination between these systems ensures high photosynthetic efficiency, but quantifying this interplay requires specific, well-defined performance benchmarks.
Establishing robust metrics is fundamental for diagnosing experimental outcomes, troubleshooting coordination failures, and guiding bioengineering efforts. This technical support center provides a foundational framework of protocols, troubleshooting guides, and reagent solutions to standardize the assessment of CCM performance in algal systems.
Key Performance Benchmarks and Quantitative Metrics
The table below summarizes the core quantitative metrics used to evaluate the performance and contribution of biophysical and biochemical CCMs.
Table 1: Core Performance Metrics for CCM Coordination
| Metric Category | Specific Metric | Experimental Assay/Method | Interpretation & Benchmark |
|---|---|---|---|
| Photosynthetic Efficiency | Ci-Specific Photosynthetic O2 Evolution Rate [2] | Clark-type O2 electrode under controlled Ci conditions [2] | Higher rates under limiting CO2 indicate a more effective CCM. A decline upon inhibitor application indicates the contribution of the targeted CCM. |
| Ci Affinity (Km(Ci)) [56] | O2 evolution or CO2 fixation assays across a Ci gradient | A lower Km value indicates a higher affinity for Ci and a more efficient CCM. | |
| Enzymatic Activity | Carbonic Anhydrase (CA) Activity [2] | Electrometric or spectrophotometric assay of CO2 hydration | High activity is indicative of an active biophysical CCM. Inhibition by EZ reduces Ci conversion near Rubisco. |
| C4 Enzyme Activity (PEPCK) [2] | Spectrophotometric monitoring of NADH oxidation or other coupled reactions | High activity suggests an operational biochemical CCM. Inhibition by MPA directly impairs the C4 acid decarboxylation step. | |
| CCM Contribution | Relative CCM Contribution [2] | Photosynthesis assay with specific inhibitors (e.g., EZ for biophysical, MPA for biochemical CCM) | The percentage decrease in carbon fixation upon inhibitor application reveals the relative contribution of each CCM type. Biophysical CCM can dominate, contributing ~100% in some species [2]. |
| Carbon Flux & Partitioning | Fatty Acid/Triacylglycerol (TAG) Biosynthesis [8] | Radiolabeled carbon tracing, lipid extraction, and chromatography | Impairment under pyrenoid disruption links CCM function to downstream carbon storage, serving as a benchmark for metabolic integration. |
| Subcellular Organization | Pyrenoid Integrity & Protein Localization [8] [71] | Immunofluorescence, electron microscopy | Relocalization of ACCase condensates to the pyrenoid periphery under low CO2 is a benchmark of functional CCM-metabolism coordination [8]. |
Essential Research Reagent Solutions
The following table details key reagents and their critical functions in CCM research, as identified from experimental protocols.
Table 2: Key Research Reagent Solutions for CCM Experiments
| Reagent / Tool | Function / Target | Application in CCM Research |
|---|---|---|
| Ethoxyzolamide (EZ) [2] | Inhibitor of carbonic anhydrase (CA) | Suppresses the biophysical CCM by inhibiting the interconversion of HCO3- and CO2, allowing quantification of its contribution to carbon fixation. |
| 3-Mercaptopicolinic Acid (MPA) [2] | Inhibitor of phosphoenolpyruvate carboxykinase (PEPCK) | Suppresses the biochemical CCM by blocking the decarboxylation of C4 acids, enabling measurement of its role in photosynthesis. |
| Acetazolamide (AZ) [2] | Specific inhibitor of external/periplasmic CA | Selectively inhibits extracellular CA activity, used to dissect internal vs. external components of the biophysical CCM. |
| Anti-ACCase Antibodies [8] | Labeling acetyl-CoA carboxylase subunits | Immunofluorescence localization to monitor the dynamic, condition-dependent formation of protein condensates at the pyrenoid periphery. |
| Anti-LCIB Antibodies [8] | Labeling the carbonic anhydrase LCIB | Visualizes the pyrenoid's outer layer, a critical microdomain for bicarbonate generation and CCM-metabolism crosstalk. |
Frequently Asked Questions (FAQs) & Troubleshooting
Q1: In our inhibitor experiments with Ulva prolifera, application of EZ (CA inhibitor) caused a ~50% drop in carbon fixation. What does this mean, and what should we check next?
- Interpretation: This indicates that the biophysical CCM contributes approximately 50% to the total carbon fixation under your test conditions. The remaining fixation is likely supported by the biochemical CCM or other pathways.
- Troubleshooting Steps:
- Verify Inhibitor Specificity: Confirm the EZ concentration was sufficient to fully inhibit CA activity without off-target effects. A dose-response curve can validate this.
- Probe the Biochemical CCM: Apply MPA (PEPCK inhibitor) alone. If fixation drops, it confirms a functional biochemical CCM. A combined EZ+MPA treatment should show an additive effect, potentially reducing fixation close to zero.
- Check Environmental Conditions: The relative contribution of each CCM is plastic. Re-run the experiment under different light intensities, Ci levels, or stress conditions, as this can shift the balance between mechanisms [2].
Q2: We are engineering a pyrenoid-deficient mutant of Chlamydomonas. What key performance benchmarks should we measure to confirm a dysfunctional CCM?
- Expected Phenotype: Pyrenoid disruption directly impairs the core biophysical CCM, leading to poor growth under low CO2 and increased photorespiration.
- Required Benchmarks:
- Ci Affinity: Measure the Km(Ci) for photosynthesis. Mutants will show a significantly higher Km (lower affinity) compared to wild-type.
- Carbon Utilization: Conduct metabolic tracing. You should observe impaired incorporation of 14C into fatty acids and triacylglycerols (TAGs), as the pyrenoid is crucial for supplying carbon to FAS [8].
- Compensatory Mechanisms: Monitor the expression and activity of C4 cycle enzymes (PEPC, PEPCK). The mutant may upregulate the biochemical CCM as a compensatory response.
Q3: We observed pyrenoid formation in our Chlamydomonas reinhardtii cultures even under high CO2 conditions. What could be the cause?
- Potential Cause: Pyrenoid induction is not exclusively linked to low CO2. Recent research shows that hyperoxia (high O2 levels) can also induce pyrenoid formation, even when CO2 is abundant [71].
- Investigation Protocol:
- Measure Dissolved Oxygen: Monitor O2 levels in your culture medium. High-density cultures in sealed bioreactors can quickly become hyperoxic due to active photosynthesis.
- Test the Hydrogen Peroxide Hypothesis: The hyperoxia signal is likely mediated by hydrogen peroxide (H2O2). Correlate pyrenoid formation with intracellular H2O2 levels using specific dyes or probes [71].
- Assess Pyrenoid Functionality: Check if the hyperoxia-induced pyrenoids are functional by testing the algae's affinity for Ci and its growth rate under low CO2.
Q4: What is the most direct evidence for a functional biochemical (C4-type) CCM in a green alga?
- Minimum Evidence Required: The simultaneous presence of three factors is considered strong evidence:
- Key C4 Enzymes: Detection of significant activity of C4-pathway enzymes like PEPC and PEPCK.
- C4 Acid Formation & Flux: Direct measurement of the formation of C4 acids (e.g., malate, aspartate) upon Ci fixation and the subsequent flux of carbon from these acids into the Calvin cycle, typically using 14C or 13C isotopic tracing.
- Physiological Impact: A measurable decrease in photosynthetic carbon fixation upon inhibition of a key C4 enzyme (e.g., with MPA for PEPCK) [2].
Experimental Protocols & Workflows
Protocol 1: Assessing Relative CCM Contributions Using Pharmacological Inhibition
This protocol allows for the dissection of the relative contributions of biophysical and biochemical CCMs to photosynthetic carbon fixation.
Detailed Methodology [2]:
- Culture and Acclimation: Grow algal cultures (e.g., Ulva prolifera) under standard conditions to a mid-log phase. Acclimate them to the specific experimental conditions (e.g., light, temperature, Ci level) for at least 24 hours.
- Ci Depletion: Cut algal fragments and transfer them to a buffered artificial seawater medium (e.g., 20 mM Hepes-NaOH, pH 8.0). Aerate the medium at low pH with pure N2 to remove CO2 and incubate for 30 minutes to deplete endogenous Ci stores.
- Inhibitor Application: Prepare separate reaction vessels with the buffered medium. Add inhibitors to final concentrations: 50 µM EZ for CA inhibition, 1.5 mM MPA for PEPCK inhibition, a combination of both, and a control with no inhibitor.
- Initiate Reaction: Add 2 mM NaHCO3 as the Ci source to each vessel.
- Measure Photosynthesis: Immediately measure the rate of photosynthetic O2 evolution using a Clark-type O2 electrode system at a controlled temperature and saturating quantum irradiance (e.g., 200 µmol photons m-2 s-1).
- Data Analysis: Calculate the percentage inhibition for each treatment. The drop in the O2 evolution rate relative to the control directly indicates the contribution of the inhibited CCM to total carbon fixation.
Protocol 2: Analyzing Pyrenoid-Associated Metabolic Coordination
This protocol investigates the dynamic localization of metabolic enzymes to the pyrenoid as a benchmark of CCM-metabolism coordination.
Detailed Methodology [8]:
- Conditional Treatment: Grow Chlamydomonas reinhardtii in two parallel environments: one bubbled with high CO2 (e.g., 5%) and another with ambient air or low CO2 (0.04%) for a minimum of 4-6 hours to fully induce the CCM.
- Cell Fixation and Processing: Harvest cells by gentle centrifugation and fix them with a suitable fixative (e.g., 4% formaldehyde for immunofluorescence). For electron microscopy, follow standard protocols for embedding and ultrathin sectioning.
- Immunolabeling: Permeabilize fixed cells and incubate with primary antibodies specific to your proteins of interest (e.g., subunits of acetyl-CoA carboxylase/ACCase for fatty acid biosynthesis, LCIB for the pyrenoid layer, Rubisco for the pyrenoid core). Subsequently, incubate with fluorescently conjugated secondary antibodies for confocal microscopy or gold-conjugated antibodies for EM.
- Imaging and Analysis: Capture high-resolution images using confocal microscopy or transmission electron microscopy. Analyze the images for protein localization patterns. A key benchmark is the redistribution of ACCase from a diffuse stromal pattern under high CO2 to concentrated condensates at the pyrenoid periphery under low CO2 [8].
- Functional Correlation: In parallel experiments, quantify the fatty acid and triacylglycerol content of the cells from both conditions using lipid extraction and chromatographic methods. Pyrenoid-deficient mutants should show impaired TAG biosynthesis under CCM-inducing conditions, linking structure to function [8].
Conclusion
The coordinated operation of biophysical and biochemical CCMs represents a sophisticated adaptive strategy that enhances algal photosynthetic efficiency under dynamic environmental conditions. Research demonstrates these mechanisms operate complementarily, with biophysical CCMs typically dominating carbon fixation while biochemical pathways provide crucial support and compensation when needed. The development of advanced methodologies, from precise inhibitor studies to synthetic encapsulin systems, has enabled unprecedented insight into CCM regulation and coordination. Future directions should focus on harnessing this knowledge to engineer optimized CCM systems in both native and heterologous hosts, with significant implications for enhancing carbon sequestration, biofuel production, and potentially informing novel carbon-capture technologies for biomedical applications. The translation of efficient algal carbon concentration strategies into other biological systems represents a promising frontier for addressing global challenges in energy, climate, and sustainable manufacturing.
References
- 1. Biophysical carbon concentrating mechanisms in land plants [https://pmc.ncbi.nlm.nih.gov/articles/PMC10802268/]
- 2. The contribution of biophysical and biochemical CO2 ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12413351/]
- 3. Response to the CO2 concentrating mechanisms and ... [https://www.sciencedirect.com/science/article/abs/pii/S1568988324001604]
- 4. Diversity of CO2 Concentrating Mechanisms in Macroalgae ... [https://www.mdpi.com/2077-1312/11/10/1911]
- 5. Introducing an algal carbon-concentrating mechanism into higher plants: location and incorporation of key components - PubMed [https://pubmed.ncbi.nlm.nih.gov/26538195/]
- 6. The contribution of biophysical and biochemical CO 2 ... [https://ui.adsabs.harvard.edu/abs/2024MLST..tmp...82Z/abstract]
- 7. The green algae CO2 concentrating mechanism and ... [https://www.nature.com/articles/s41467-025-60525-7]
- 8. Algal carbon concentrating drives fatty acid biosynthesis ... [https://www.sciencedirect.com/science/article/pii/S2211124725012070]
- 9. Introducing an algal carbonâ€concentrating mechanism into ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5102585/]
- 10. Carbonic Anhydrase 4 - an overview [https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/carbonic-anhydrase-4]
- 11. The Role of the C4 Pathway in Carbon Accumulation and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC520782/]
- 12. Role of C4 carbon fixation in Ulva prolifera, the macroalga ... [https://www.nature.com/articles/s42003-020-01225-4]
- 13. Energy crosstalk between photosynthesis and the algal ... [https://www.sciencedirect.com/science/article/abs/pii/S1360138523000985]
- 14. The Prospect of Using Cyanobacterial Bicarbonate ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3075752/]
- 15. SLC4A Transporters - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4768801/]
- 16. The pyrenoid: the eukaryotic CO2-concentrating organelle [https://pmc.ncbi.nlm.nih.gov/articles/PMC10473226/]
- 17. Pyrenoid [https://en.wikipedia.org/wiki/Pyrenoid]
- 18. Modelling the pyrenoid-based CO2-concentrating ... [https://www.nature.com/articles/s41477-022-01153-7]
- 19. A pyrenoid-based CO2-concentrating mechanism can ... - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12343021/]
- 20. Transcriptomic and proteomic responses to very low CO2 ... [https://biotechnologyforbiofuels.biomedcentral.com/articles/10.1186/s13068-019-1506-8]
- 21. Machine learning optimization of environmental factors ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12040456/]
- 22. Frequently asked questions about in vivo chlorophyll ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4210649/]
- 23. Chlorophyll fluorescence [https://en.wikipedia.org/wiki/Chlorophyll_fluorescence]
- 24. Impact of nutrients on photoacclimation of phytoplankton in an... [https://link.springer.com/article/10.1007/s10750-020-04213-1]
- 25. Optimizing Growth Conditions and Biochemical Properties ... [https://www.mdpi.com/2076-3417/15/2/810]
- 26. Chlorophyll content and chlorophyll fluorescence analysis [https://bio-protocol.org/exchange/minidetail?id=6990379&type=30]
- 27. Stability-activity tradeoffs constrain the adaptive evolution ... [https://pubmed.ncbi.nlm.nih.gov/24469821/]
- 28. Investigating Photosynthesis – Biology Laboratory Manual [https://iastate.pressbooks.pub/isudp-2025-205/chapter/photosynthesis/]
- 29. Metabolomics and isotope tracing - PMC - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC6034115/]
- 30. A Beginner's Guide to Metabolic Tracing [https://bitesizebio.com/81841/metabolic-tracing-guide/]
- 31. Troubleshooting in Large-Scale LC-ToF-MS Metabolomics ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6918290/]
- 32. Global stable-isotope tracing metabolomics reveals system ... [https://www.nature.com/articles/s41467-022-31268-6]
- 33. Guide to Metabolomics Analysis: A Bioinformatics Workflow [https://pmc.ncbi.nlm.nih.gov/articles/PMC9032224/]
- 34. Optimizing biofuel production from algae using four- ... [https://www.sciencedirect.com/science/article/pii/S2352484724004153]
- 35. Advancing ecological and environmental biology research [https://pmc.ncbi.nlm.nih.gov/articles/PMC12413372/]
- 36. Beyond Cutting: CRISPR-Driven Synthetic Biology Toolkit for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12347677/]
- 37. Genetic Engineering of Algae for Enhanced Biofuel Production [https://pmc.ncbi.nlm.nih.gov/articles/PMC2863401/]
- 38. Biocontainment of Genetically Engineered Algae [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2022.839446/full]
- 39. Multi-omics integrative analysis reveals novel genetic loci and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12328896/]
- 40. Collaboration Across Omics: Integrating Spatial Proteomic ... [https://www.syncell.com/spatial-proteomics-blog/collaboration-across-omics-integrating-spatial-proteomic-discovery-with-genomics-and-transcriptomics/]
- 41. Enhancing CO2 fixation by microalgae in a Photobioreactor [https://www.sciencedirect.com/science/article/abs/pii/S0960852424008800]
- 42. The contribution of biophysical and biochemical CO2 ... [https://pubmed.ncbi.nlm.nih.gov/40919461/]
- 43. Reprogramming encapsulins into modular carbon-fixing ... [https://www.nature.com/articles/s41467-025-65307-9]
- 44. Synthetic biology to supercharge photosynthesis in crops [https://www.eurekalert.org/news-releases/1103768]
- 45. Synthetic biology to supercharge photosynthesis in crops [https://www.sydney.edu.au/news-opinion/news/2025/11/04/synthetic-biology-encapsulins-photosynthesis-szyszka.html]
- 46. Reprogramming encapsulins into modular carbon-fixing ... [https://pubmed.ncbi.nlm.nih.gov/41168233/]
- 47. Light-independent regulation of algal photoprotection by ... [https://www.nature.com/articles/s41467-023-37800-6]
- 48. Navigating microalgae under dynamic CO2 for waste-to- ... [https://www.sciencedirect.com/science/article/abs/pii/S016777992500229X]
- 49. High-value biomass from microalgae production platforms [https://pmc.ncbi.nlm.nih.gov/articles/PMC6100726/]
- 50. Advances in the optimization of central carbon metabolism in ... [https://microbialcellfactories.biomedcentral.com/articles/10.1186/s12934-023-02090-6]
- 51. Promising solutions to solve the bottlenecks in the large- ... [https://www.sciencedirect.com/science/article/abs/pii/S1364032116002161]
- 52. Mitochondria Affect Photosynthetic Electron Transport and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5841685/]
- 53. Chloroplast pH Homeostasis for the Regulation of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9174948/]
- 54. Optimal energy and redox metabolism in the ... [https://www.nature.com/articles/s41540-023-00307-3]
- 55. Potential and Challenges of Improving Photosynthesis in ... [https://www.mdpi.com/2223-7747/9/1/67]
- 56. Adapting from Low to High: An Update to CO2-Concentrating ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10096703/]
- 57. A Reliable and Non-destructive Method for Monitoring the ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2017.02079/full]
- 58. The green algae CO2 concentrating mechanism and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12174325/]
- 59. 绿藻的CO浓缩机制和光呼å¸åœ¨é€‚应低CO环境的过程ä¸å…±åŒ ... [https://suppr.wilddata.cn/browses/detail/pubmed/40527898]
- 60. 光呼å¸æ¼”化ã€è°ƒæŽ§ä¸Žé—ä¼ æ”¹è‰¯ [https://lifescience.sinh.ac.cn/202410/20241002.htm]
- 61. Photosynthetic efficiency and transcriptome analysis of ... [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2023.1192258/full]
- 62. Enhancing Photosynthesis and Plant Productivity through ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11352682/]
- 63. Will an algal CO2-concentrating mechanism work in higher ... [https://www.sciencedirect.com/science/article/abs/pii/S1369526616300668]
- 64. Optimization and Process Effect for Microalgae Carbon ... [https://www.mdpi.com/2311-5629/9/1/35]
- 65. Measuring excitation-inhibition balance through spectral ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10849740/]
- 66. Modeling with uncertainty quantification reveals the essentials ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11836721/]
- 67. Enhancing carbon capture and lipid accumulation by ... [https://www.sciencedirect.com/science/article/abs/pii/S1876107018305674]
- 68. Cultivation modes affect the morphology, biochemical ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11511745/]
- 69. Advancing the scale of synthetic biology via cross-species ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10943186/]
- 70. Beyond Cutting: CRISPR-Driven Synthetic Biology Toolkit ... [https://www.mdpi.com/1422-0067/26/15/7470]
- 71. Photosynthesis: Oxygen overload [https://prl.natsci.msu.edu/news-and-events/news/photosynthesis-oxygen-overload.aspx]